Genetic Characterization of Second-Line Drug-Resistant and Extensively Drug-Resistant Mycobacterium tuberculosis from the Northern Region of India
- DOI
- 10.2991/j.jegh.2018.02.100How to use a DOI?
- Keywords
- Tuberculosis; cross resistance; mutation; MAS PCR; DNA sequencing
- Abstract
Rapid detection of drug resistance in Mycobacterium tuberculosis is important for the successful treatment of tuberculosis. Fluoroquinolone and aminoglycoside resistance detection by molecular methods becomes more complex due to cross resistance among them. Thus, we aimed to determine cross-resistance and mutations in resistance genes for these drugs. A total of 336 multidrug-resistant tuberculosis (MDR-TB) cases received in Mycobacteriology laboratory were screened for phenotypic drug sensitivity testing for second-line drugs, i.e., ofloxacin, amikacin, kanamycin, and capreomycin. Molecular characterization of resistance was done by DNA sequencing of gyrA gene for fluoroquinolones (FQ), and multiplex allele-specific polymerase chain reaction (PCR) and PCR restriction fragment length polymorphism (RFLP) of rrs gene for aminoglycosides. Of 336 MDR-TB isolates, 12 were extensively drug-resistant tuberculosis and 219 were sensitive to all the drugs tested. Ofloxacin, amikacin, kanamycin, and capreomycin resistance was detected in 101 (30.1%), 23 (6.8%), 27 (8.1%), and 19 (5.6%) cases, respectively. Eight different mutations were detected in gyrA gene in ofloxacin-resistant isolates and A1401G nucleotide change in rrs gene were seen in 55.6% (15/27), 65.2% (15/23), and 68.4% (13/29) for kanamycin-, amikacin-, and capreomycin-resistant isolates, respectively. Information on second-line drug resistance-associated mutations could potentially be used for development of newer rapid diagnostic tests.
- Copyright
- © 2018 Atlantis Press International B.V.
- Open Access
- This is an open access article under the CC BY-NC license (http://creativecommons.org/licences/by-nc/4.0/).
1. INTRODUCTION
Drug-resistant tuberculosis is a major public health problem in developing countries, which also threatens the success of tuberculosis (TB) control programs at the national level. An estimated 300,000 pulmonary multidrug-resistant-TB (MDR-TB) patients were notified in 2014, and more than half of these patients were in India, China, and the Russian Federation. Globally, the success rate of treatment of MDR-TB is only 50%, largely due to high rates of mortality and loss to follow-up. Among the MDR-TB patients, an estimated 9.7% of people have extensively drug-resistant tuberculosis (XDR-TB) [1]. MDR-TB is defined as Mycobacterium tuberculosis strain resistant to at least rifampin and isoniazid, and XDR-TB is MDR M. tuberculosis and also resistant to one of the second-line injectable drugs (SCID; amikacin, kanamycin, or capreomycin) and a fluoroquinolone. Rapid detection of MDR- and XDR-TB is necessary for the selection of correct treatment regimen and to reduce the transmission of drug-resistant strains [2].
The XDR-TB diagnosis is much more complex than that of MDR-TB due to the absence of rapid and good molecular diagnostic methods. Phenotypic drug sensitivity testing (DST) for second-line drugs, i.e., ofloxacin, levofloxacin, moxifloxacin, and gatifloxacin (fluoroquinolones) and kanamycin, amikacin, and capreomycin (injectable drugs) is reliable and reproducible across various settings. However, the turnaround time of phenotypic DST is in terms of weeks because of the slow growth rate of M. tuberculosis. The development of rapid molecular diagnostic test for the detection of second-line resistance needs an accurate knowledge of mutations in the drug-targeted genes from different geographical regions. Molecular diagnosis promises rapid detection of drug resistance, which may be critical in high-burden areas of MDR- and XDR-TB to break the transmission and development of new drug resistance. The molecular mechanisms of drug resistance to second-line antituberculosis drugs have been elucidated [3]. Fluoroquinolones (FQ) resistance in M. tuberculosis is mainly due to mutations in quinolone resistance-determining region (QRDR), which is a short nucleotide sequence of the gene gyrA and, less frequently, in gyrB. The majority of mutations of gyrA gene have been correlated with FQ resistance in clinical isolates of M. tuberculosis. Resistance to SCID is mainly associated with point mutation in the 16S rRNA gene (rrs) at the 1401 position [4,5].
The available molecular methods cannot replace phenotypic DST for second-line antibiotics because there is an incomplete cross-resistance among second-line drugs. The members of different groups of second-line drugs share common mutations in their targeted region, thus making it problematic to differentiate between the cross-resistance among those members. The occurrence of mutations in the targeted regions also varies according to the geographical region. The data from limited studies showed that up to 87% of fluoroquinolone and up to 70% of second-line aminoglycoside resistance is caused by point mutation within the gyrA and rrs gene, respectively [6]. So, this study was planned to understand the cross-resistance and genetically characterize the isolates of this geographical area.
2. MATERIALS AND METHODS
2.1. M. tuberculosis Isolates and Drug Susceptibility Testing
Three hundred and thirty-six (336) pulmonary MDR-TB isolates, which were received between September 2015 to June 2016, were taken from cultures and drug sensitivity testing laboratory, Mycobacteriology Section, Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research, Chandigarh (An RNTCP approved center for TB culture and DST). These isolates were received for routine second-line drug testing of M. tuberculosis under the RNTCP program. M. tuberculosis isolates were subcultured and tested for second-line drug sensitivity in BACTEC MGIT 960 (BD Bioscience, Belgium) according to the manufacturer’s instructions (Becton and Dickinson, USA). M. tuberculosis isolates were subjected to second-line drug sensitivity testing using World Health Organization (WHO)-recommended critical concentrations [7] i.e., ofloxacin, 2.0 mg/L; amikacin, 1.0 mg/L; capreomycin, 2.5 mg/L; and kanamycin, 2.5 mg/L (Sigma-Aldrich, USA). Because the observations were made as a part of the national TB control program, a separate ethical clearance was not required.
2.2. Genomic DNA Extraction
One milliliter culture from MGIT was centrifuged, and the pelleted cells were resuspended in 500 μl of distilled H2O and heat inactivated at 95°C for 10 min. Genomic DNA was extracted using a modified cetyltrimethylammonium bromide–sodium chloride method [8].
2.3. PCR Amplification and PCR RFLP
Polymerase chain reaction (PCR) was performed to amplify the partial gyrA and rrs genes of M. tuberculosis. The mutations in gyrA gene were screened by 248 bp PCR amplification followed by DNA sequencing. The Multiplex Allele Specific (MAS) PCR and PCR restriction fragment length polymorphism (RFLP) were used for the detection of A1401G mutation in rrs gene responsible for aminoglycoside resistance in M. tuberculosis [9]. PCR amplification for gyrA was performed in a 25 μl reaction mixture containing 2.5 μl of 10× PCR buffer (Fermentas, Lithuania), 1.5 μl of 10 μM forward (gyr AF) and reverse (gyr AR) primers (Sigma Aldrich, India), 200 μM deoxyribonucleotide triphosphate (dNTP) (Fermentas, Lithuania), 1 U Taq DNA polymerase (Fermentas, Lithuania), and 1 μl genomic DNA. The PCR conditions were as follows: initial denaturation at 95°C for 5 min; 30 cycles including denaturation at 95°C for 45 s, annealing at 58°C for 45 s, and extension at 72°C for 45 s; and last, a final extension at 72°C for 6 min. MAS PCR assay was performed to identify rrs A1401G mutation. PCR assays had a final volume of 25 μl and contained 2.5 μl of 10× PCR buffer (Fermentas, Lithuania), 1.5 U of Taq DNA polymerase (Fermentas, Lithuania), and 50–200 ng of genomic DNA as template. For amplification of rrs, 200 μM of dNTPs (Fermentas, Lithuania), 20 pmol of forward primer RRSF, 5 pmol of internal reverse primer RRSR1, and 40 pmol of reverse primer RRSR were used. For PCR RFLP, the same primers RRSF and RRSR were used followed by the restriction digestion with BmgBI (New England Biolabs, UK). Fragments of 353 and 128 bp were obtained from PCR RFLP of rrs gene showing the wild-type genotype (Fig. 1).
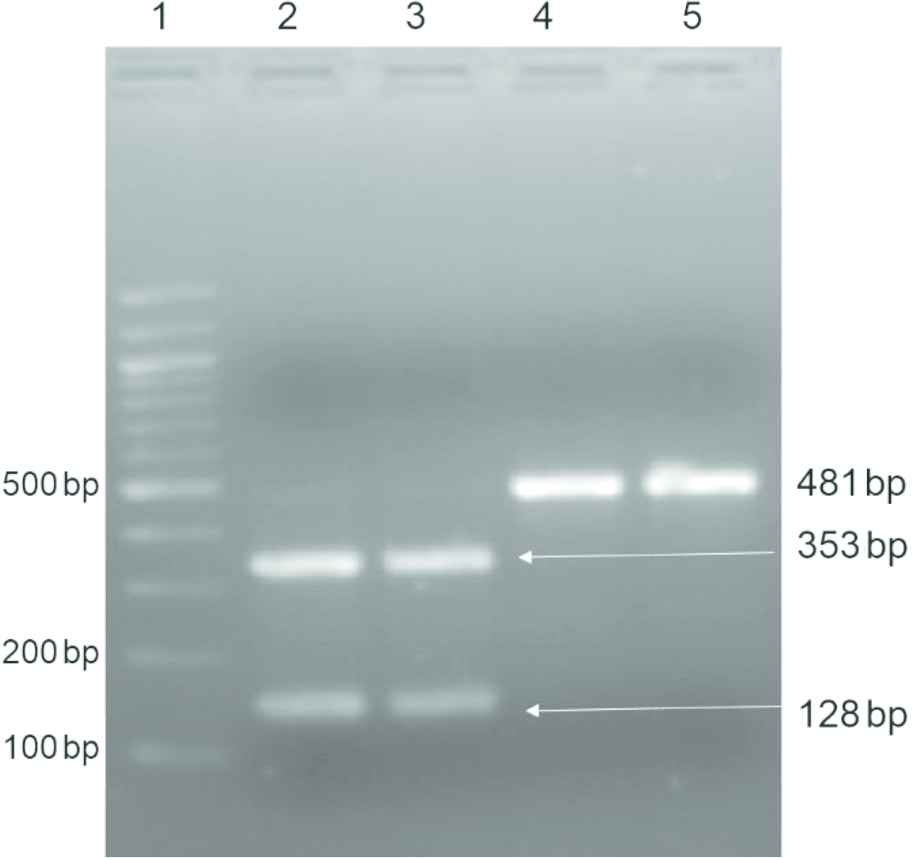
PCR RFLP of rrs gene after digestion with BmgBI. Lane 1, 100 bp DNA ladder; Lanes 2 and 3, 353 and 128 bp fragments with wild-type sequence; and Lanes 4 and 5, 481 bp fragments of mutant sequence
2.4. DNA Sequencing
The gene gyrA of all ofloxacin resistant isolates and the representative samples of rrs gene (5 resistant isolates) were sequenced for mutation analysis. Sequence reactions were carried out in ABI 3130 Genetic Analyzer automated sequencer (Applied Biosystems, USA) following the manufacturer’s instructions. Sequencing primers were identical to the primers used for PCR amplification.
3. RESULTS
3.1. Drug-resistance Patterns among the Clinical Isolates
Among the 336 cases, 215 (63.9%) were male and 121 (36%) were female, and their ages ranged from 12 to 72 years. DST revealed that 229 (68.1%) isolates were susceptible in vitro to all the second-line drugs tested; and 107 (31.8%) were resistant to any of the drugs tested. Of the 107 drug-resistant isolates, 12 (3.6%) were found to be XDR-TB. Overall, 89 (26.5%) isolates were mono-resistant to ofloxacin and none to kanamycin, amikacin, and capreomycin. Any resistance to ofloxacin, kanamycin, amikacin, and capreomycin was found in 101 (30.1%), 27 (8%), 23 (6.8%), and 19 (5.6%) isolates, respectively. Among 12 (3.6%) XDR-TB isolates, three were resistant to ofloxacin and kanamycin and another three were resistant to ofloxacin, kanamycin, amikacin, and capreomycin. Four isolates were resistant to ofloxacin, amikacin, and kanamycin, one was resistant to ofloxacin, amikacin, and capreomycin, and yet another isolate was resistant to ofloxacin, kanamycin, and capreomycin.
3.2. Cross-resistance of Isolates to SCID
Among the 28 (8.3%) SCID-resistant isolates, kanamycin, amikacin, and capreomycin resistance was found in 27 (8.1%), 23 (6.8%), and 19 (5.6%) isolates, respectively. The cross-resistance among the SCID was found in 25 isolates except in three, which were only resistant to kanamycin. The cross-resistance between three aminoglycosides, i.e., kanamycin, amikacin, and capreomycin was found in 16 isolates and between kanamycin and amikacin in six isolates. Kanamycin and capreomycin or amikacin and capreomycin cross-resistance were found in two and one cases, respectively (Fig. 2).
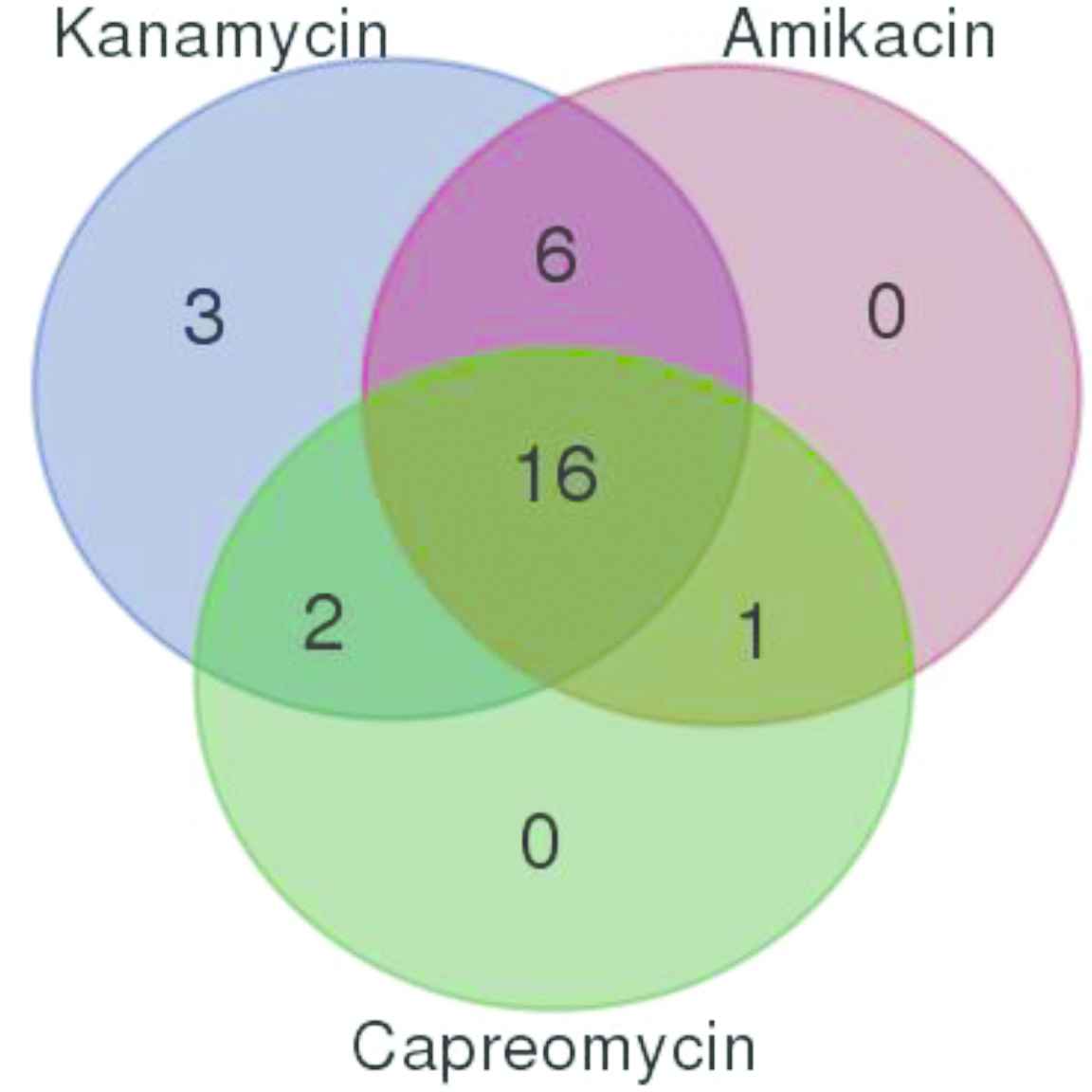
Cross-resistance of MDR TB isolates to second-line injectable drugs
3.3. Molecular Characterization of Drug Resistance
3.3.1. Ofloxacin resistance
A total of 101 ofloxacin resistant isolates (12 XDR-TB and 89 mono-resistant) were tested for mutations in the partial gyrA gene, including the core region QRDR. The 248 bp amplified PCR product was sequenced, and analysis showed that 92 (91.1%) isolates had a mutation in the gyrA gene and nine (8.9%) isolates did not show any mutation in the target region (Table 1). Codon 94 was found to be the most frequently mutated codon in 55 (54.5%) ofloxacin-resistant isolates. At codon 94, five types of changes were observed: Asp94Gly was found in 34, Asp94Ala in nine, Asp94Asn in six, Asp94Tyr in three, and Asp94His was found in three isolates. At codon 90, only one type of change, i.e., Ala90Val was detected in 24 (23.7%) isolates. Ala90Val/Ser91Pro double change was found in nine isolates, whereas a single change on codon 91 (Ser91Pro) was found in four isolates (Table 1).
| Drug | Phenotype (No.) | No | Mutation | Codons |
|---|---|---|---|---|
| Ofloxacin | Resistant (101) | 34 | Asp94Gly | GAC 94 GGC |
| 9 | Asp94Ala | GAC 94 GCC | ||
| 6 | Asp94Asn | GAC 94 AAC | ||
| 3 | Asp94Tyr | GAC 94 TAC | ||
| 3 | Asp94His | GAC 94 CAC | ||
| 24 | Ala90Val | GCG 90 GTG | ||
| 9 | Ala90Val/Ser91Pro | GCG 90 GTG/TCG 91 CCG | ||
| 4 | Ser91Pro | TCG 91 CCG | ||
| 9 | Wild Type | |||
| Kanamycin | Resistant (27) | 15 | A1401G | A1401G |
| 12 | Wild Type | |||
| Amikacin | Resistant (23) | 15 | A1401G | A1401G |
| 8 | Wild Type | |||
| Capreomycin | Resistant (19) | 13 | A1401G | A1401G |
| 6 | Wild Type |
Mutation profiles of resistant genes gyrA (ofloxacin) and rrs (kanamycin, amikacin, and capreomycin) for resistant isolates
3.3.2. SCID resistance
The rrs gene was screened by MAS PCR and confirmed by PCR RFLP for the detection of mutation A1401G. The MAS PCR and PCR RFLP showing double band at 128 and 353 bp were taken as sensitive and single band at 481 resulted in a resistant isolate (Fig. 1). The representative strains were sequenced for the confirmation of results. Of the 27 kanamycin-resistant isolates, 15 (55.6%) had A1401G rrs mutation. The A1401G rrs gene mutation were found in 15/23 (65.2%) amikacin-resistant strains and 13/19 (68.4%) capreomycin-resistant strains (Table 1). All the sensitive strains were also screened for the A1401G mutation, but none of the kanamycin- and amikacin-sensitive strains were found to be mutated at the 1401 codon. Two capreomycin-sensitive strains resistant to amikacin and kanamycin had mutations at the 1401 codon.
4. DISCUSSION
Rapid detection of drug resistance by molecular methods could be of great value in aiding the timely administration of appropriate antibiotics to MDR-TB/XDR-TB patients. In this study, drug resistance pattern of second-line antibiotics and mutations in their targets were characterized in MDR-TB isolates from North India. Among the second-line antibiotics, the molecular mechanism of drug resistance is the most completely understood for fluoroquinolone as compared with that of others. Fluoroquinolones resistance in M. tuberculosis has been predominantly attributed to gyrA mutations [5,10]. In our study, the phenotypic and genotypic concordance was found to be 91.1% for ofloxacin resistance. Of the ofloxacin-resistant isolates, 92 (91.1%) had mutations in the QRDR region of the gyrA gene, whereas nine (8.9%) had none. Mutations at gyrA codons 90, 91, or 94 were the most common in previous studies [3–5,11], and we found that 79 (78.2%) isolates had point mutations at two major codons, i.e., 94 and 90 of the core QRDR of the gyrA gene. The most frequent mutation found was Asp94Gly in 34 (33.7%) isolates, followed by Ala90Val in 24 (23.7%). Double mutation GCG-GTG/GAC-GGC was detected in nine (8.9%) isolates. Previous studies reporting mutations have shown that in Indian isolates the most common mutation of the gyrA gene is at codon 94, followed by codons 90 and 91 [12,13]. In this study, however, we found no isolates harboring mutations at codon 88, but nine isolates were detected with a double mutation in the QRDR region. A rare mutation Asp94His was detected in three isolates that was resistant to ofloxacin.
For SCID, the rrs A1401G single nucleotide polymorphism has been considered a sensitive marker for resistance, because it accounted for high-level resistance to the various drugs mentioned. Approximately 30–90% of aminoglycoside-resistant strains harbor this mutation; thus, it can be used as a surrogate marker for the detection of resistance to amikacin and kanamycin [9,14,15]. The other mutations in rrs gene are poor markers due to their presence in susceptible strains [16]. In our study, rrs mutations in aminoglycoside-resistant isolates were more common specifically at the residue 1401. The nucleotide change in rrs gene was seen in 55.6% (15/27), 65.2% (15/23), and 68.4% (13/29) for kanamycin-, amikacin-, and capreomycin-resistant strains, respectively. No kanamycin- and amikacin-sensitive strains had mutations on this site. Two capreomycin-sensitive isolates had A1401G mutation suggesting <100% specificity for the prediction of capreomycin-resistant phenotypes.
Treatment outcome of MDR-TB cases is poor because it relies on second-line drugs that are less-potent and more toxic, and owing to the emergence of drug resistance. For better management of MDR-TB, we need to check the activity of new compounds that can give better results [17,18]. To increase the sensitivity and specificity of molecular diagnostics for the detection of XDR-TB, we need an understanding of the frequency of different mutations in the drug-targeted region and their geographical distribution. This study characterizes mutations conferring resistance to fluoroquinolones and SCID at the common target loci in M. tuberculosis isolates from North India, and enhances our understanding of the geographic distribution of resistant alleles from this region. Such data may be valuable for the development of newer, rapid, and reliable molecular approaches for the detection of XDR-TB in clinical isolates of M. tuberculosis.
ACKNOWLEDGMENTS
The financial, administrative, and technical support of the Department of Medical Microbiology, PGIMER, Chandigarh and Revised National Tuberculosis Control Program is acknowledged.
CONFLICTS OF INTEREST
All authors declared no conflicts of interest.
REFERENCES
Cite this article
TY - JOUR AU - Rakesh Yadav AU - Aastha Saini AU - Jureka Mankotia AU - Rajiv Khaneja AU - Priyanka Agarwal AU - Sunil Sethi PY - 2018 DA - 2018/12/31 TI - Genetic Characterization of Second-Line Drug-Resistant and Extensively Drug-Resistant Mycobacterium tuberculosis from the Northern Region of India JO - Journal of Epidemiology and Global Health SP - 220 EP - 224 VL - 8 IS - 3-4 SN - 2210-6014 UR - https://doi.org/10.2991/j.jegh.2018.02.100 DO - 10.2991/j.jegh.2018.02.100 ID - Yadav2018 ER -
