Case-Based Decision Support System for Breast Cancer Management
- DOI
- 10.2991/ijcis.2018.25905180How to use a DOI?
- Keywords
- knowledge intensive case-based reasoning; breast cancer; ontology; case-based decision support system; jColibri
- Abstract
Breast cancer is identified as the most common type of cancer in women worldwide with 1.6 million women around the world diagnosed every year. This prompts many active areas of research in identifying better ways to prevent, detect, and treat breast cancer. DESIREE is a European Union funded project, which aims at developing a web-based software ecosystem for the multidisciplinary management of primary breast cancer. The development of an intelligent clinical decision support system offering various modalities of decision support is one of the key objectives of the project. This paper explores case-based reasoning as a problem solving paradigm and discusses the use of an explicit domain knowledge ontology in the development of a knowledge-intensive case-based decision support system for breast cancer management.
- Copyright
- © 2018, the Authors. Published by Atlantis Press.
- Open Access
- This is an open access article under the CC BY-NC license (http://creativecommons.org/licences/by-nc/4.0/).
1. Introduction
Breast cancer is the most common type of cancer in women worldwide, with the mortality rate being second highest (next to lung cancer) among different types of cancer.1 As reported by World Health Organization (WHO, 2013),2 508,000 women died due to breast cancer in 2011. Perceived as a disease of the developed world, incidence rates vary worldwide, from 19.3, 40, to 89.7 per 100,000 women in Eastern Africa, most of the developing countries, and Western Europe respectively.2 Whereas, the survival rates vary from 40%, 60%, to 80% in low-income, middle-income, and developed countries respectively, which reflects the lack of early detection programs and adequate diagnosis and treatment facilities in the less-developed countries.
In light of this, both industry and academia are taking mitigating action in providing clinical support solutions and patient-centric healthcare systems for breast cancer diagnosis and management. For instance, the worldwide cancer research organization3 is funding 30 research projects across the world for breast cancer, among which 23 projects are researched and developed in Western European countries. In UK, various breast cancer related research projects are being funded by Cancer Research UK,4 National Breast Cancer Foundation,5 Breast Cancer UK6 etc. DESIREE is a European Union funded project,a which aims at developing a web-based software ecosystem for the personalized, collaborative, and multidisciplinary management of primary breast cancer (PBC) by multidisciplinary Breast Units (BUs). With the ultimate goal of the system to be deployed in actual BUs in the future, one of the main objectives of the project is to develop an intelligent clinical decision support system (DSS). The idea is to go beyond the limitations of clinical practice guidelines for breast cancer care,7 and incorporate experience-based and case-based decision support modalities that are based on the evolving knowledge acquired from previous patient cases. In this paper, we present the knowledge intensive case-based reasoning (KI-CBR) model, which incorporates knowledge from explicit domain knowledge we developed and serves as the case-based DSS (CB-DSS) within the DESIREE project. The proposed model conceptualizes and integrates the breast cancer domain knowledge in an ontological format and the case retrieval algorithm using semantic similarity measure.
Case-based Reasoning (CBR) has been a field of great interest for researchers, entrepreneurs, as well as clinicians for over three decades.8,9,10 It integrates several disciplines of heterogeneous nature such as Cognitive Science, Artificial Intelligence (AI), and Information Science to provide a computational model that is very close to human reasoning. The main advantage is that with CBR, the usually extensive and complex formalization of the problems to be solved is not required. Unlike various other AI approaches such as rule-based reasoning, or neural networks, that generate abstract representations from a set of training examples, CBR methodology adapts instance-based learning and uses previous similar cases as the basis for decision making. In medical domain in particular, the clinicians not only use the rules from clinical practice guidelines, but also use their knowledge and experience to diagnose the disease and provide treatment. In a CBR model, the experience can be incorporated as cases in the case-base, which could include not only typical simple scenarios, but also the complex and exceptional ones. Thus, the model considers them when reasoning and automatically enriches the case-base with parts of changeable knowledge as new case and enables it to perform better with usage. Thus, as the clinical decisions made by physician improves with experience, the performance of CBR model would also improve with usage, which is not possible in a rule-based model.11 Thus, the fact that the CBR methodology closely resembles the thought process of the clinicians, and the gradual acceptance of advanced decision support systems in clinical practice, suggest the success of CBR in medicine.12 Some of the CBR systems developed in medicine and health science domains so far, include CASEY13 to diagnose heart failure patients, MNAOMIA14 to diagnose and treat eating disorders, PROTOS15 for hearing disorder diagnosis, MacRad16 for radiology image classification, GerAmi17 for Alzheimer’s disease management, and GOCBR18 for breast cancer diagnosis. Many such CBR applications are well summarized by Choudhury et al.11
When developing a CBR system, the main challenge is to build the case base and implement an effective case retrieval algorithm. In a medical application, acquiring and managing a complete and consistent case base, covering a large number of resolved patient cases with varying diagnosis output, becomes the basis for building a reliable CB-DSS. Then, CBR would adapt a supervised learning algorithm, trained on resolved cases, stored in the case base. Thus, first identifying the most similar cases from the case base, and then adapting the solution of the retrieved cases to build the solution for the new query case, solves a new case. Taking into account of various algorithmic strategies19 such as root mean square distance, geometrical matching, consensus shapes, weighted Euclidean distance etc., in developing similarity analysis and case retrieval algorithm, which would accurately measure the clinical distance between two patient cases, becomes a part of the problem-solving paradigm. In the present work, we present an ontology-based schema to conceptualize, interpret, and integrate large clinical data of breast cancer patients into a case-base knowledge. Subsequently, benefiting from the knowledge codified in the ontology, we incorporate semantic similarity measure to implement the case retrieval algorithm.
Implementation of CBR algorithm is feasible on various platforms. However, building the CBR framework to execute the complete CBR cycle is a challenge, which requires substantial research effort and time. In light of which, with the effort to formalize CBR and provide design and implementation assistance, various CBR frameworks have been distributed as software tools. Some of the popular non-commercial tools include myCBR, jColibri, CAT-CBR, CASPAIN, CBR Shell, FreeCBR, and eXiTCBR. Based on a detailed comparative study performed on these different software tools,19,21,22 myCBR and jColibri were initially selected, and jColibri2 (Java framework)23 was finally adopted in this project to design a CB-DSS for breast cancer management.
In the following section, we present the overall framework of the proposed CB-DSS for DESIREE, describe the related functional blocks, and explain the details of the proposed CB-DSS, which include the clinical data integration using case-base knowledge model, the similarity analysis, and the case retrieval algorithm using semantic similarity measure and the patient case representation. Section 3 presents a case study to show the complete operation of the proposed CB-DSS. Finally, section 4 draws the conclusion of the paper.
2. Case-Based Decision Support System for DESIREE
2.1. Framework and Workflow
One of the main objectives of the DESIREE project is to provide decision support for the therapeutic decisions made by BUs, including surgery, radiotherapy, and systemic therapies. With the aim to provide a personalized state-of-the-art clinical decision support system to BUs, the project provides different modalities of decision support, including guideline (GL),24 experience (EX)25 and CB-DSS. In this paper, we present the proposed CB-DSS.
The DESIREE platform contains the DESIREE Information Management System (DESIMS), Fast Healthcare Interoperability Resources (HAPI-FHIR)26 server and the three DSSs, including GL-DSS, EX-DSS and CB-DSS. The DESIMS contains a clinical interface through which the clinical data of patients are entered and stored according to a common data model. The clinical interface also allows clinical partners to access the heterogeneous patient data and retrieve the results and therapeutic propositions from the different DSSs. The clinical data of patient are transferred from DESIMS to the DSSs for analysis through the HAPIFHIR server, which is an open source standard framework developed for exchanging healthcare data in a systematic manner. Fig. 1 mainly shows the framework and workflow of the CB-DSS in the DESIREE platform.
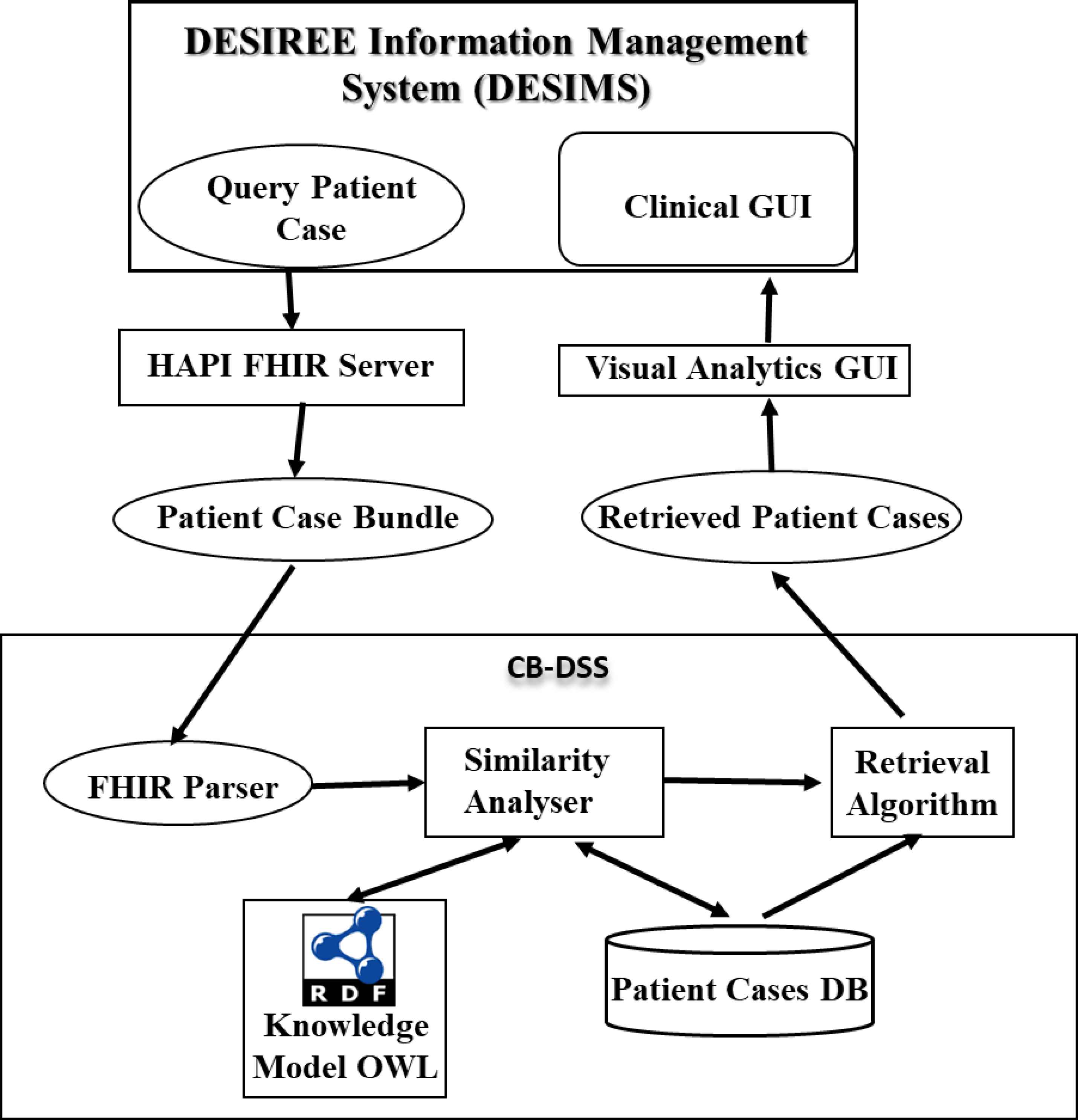
Framework and workflow of CB-DSS
In order to incorporate decisional criteria beyond the limitations of current guidelines for breast cancer management, the CB-DSS incorporates the experience of clinicians on previous cases, by collecting the description of patients, and the decision made by clinicians, as the case representation. Subsequently, it provides a tool for querying former cases in order to retrieve similar patient cases based on the defined case base and case retrieval algorithm using semantic similarity measure.
In the proposed CB-DSS shown in Fig. 1, during runtime, the CB-DSS queries the FHIR server, linked to the DESIMS, for the query patient case. Here, the query patient case represents a new patient case with a number of clinical parameters recorded by BU clinicians using the DESIMS interface. CB-DSS first parses the patient case bundle retrieved from the FHIR server as input attributes to CBR, which includes the similarity analyser and retrieval algorithm. The similarity analyser compares the query case with the patient cases stored in the database using various similarity functions, whereas, the retrieval algorithm retrieves the most similar patient cases and sends the results to the visual analytics graphical user interface (GUI). The retrieved patient cases are graphically visualized in the DESIMS by comparing the clinical parameters and the clinical decision made on retrieved cases with the information provided for the query case using the rainbow boxes visualization technique developed by Lamy et al.27
The following sections present a detailed description of the proposed CB-DSS.
2.2. Clinical Data Integration using Knowledge Model Representation
The crucial step in building any clinical DSS allies in first making sense of the large clinical information available, in terms of conceptualizing, preparing, and integrating data. In breast cancer research, relevant clinical data/information could come from various resources, including patient health records, clinical practice guidelines, medical & scientific documents, etc., which are usually in an unstructured and heterogeneous data format. Literature shows that both academia and industry have explored various approaches to data integration.28,29,30 Among which, we explore semantic technologies and use ontology, where the medical terms are organized based on concepts and the relationship between them. In particular, for CBDSS, we exploit the use of ontology-based integration to avail the advantage of the semantic similarity measure.
From the information provided by clinical partners, clinical practice guidelines, clinical documentation etc., an ontology-based breast cancer knowledge model (BCKM) was defined using the Web Ontology Language (OWL) and Protégé software tool. Although it would be ideal to use established authoritative ontologies such as the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT),b which has a greater depth of terminologies for all clinical specialties, the knowledge model was defined to exclusively embed concepts relevant to the breast cancer domain. This specifically avoided the risk of misinterpreting medical terminologies in different care settings and facilitated the analysis of different semantic similarity measures for CB-DSS. However, to establish semantic interoperability beyond the DESIREE project, attributes in the knowledge model were linked with some of the established authoritative terminologies in NCI thesaurusc and SNOMED CT.
Fig. 2 illustrates the semantic representation of the BCKM. It shows the top-level concepts, some of the relevant classes and the connections between these concepts using object properties. Fig. 3 shows a screenshot of the BCKM developed using Protégé, which highlights an attribute under the concept “Reference Concept” and its link with the standard terminology in the NCI thesaurus. It also shows the taxonomic hierarchy of the “Cancer TNM Staging”. This architecture, allows one to examine the path length, depth and local density factors associated with the ontological taxonomic hierarchy to compute the similarity measure.
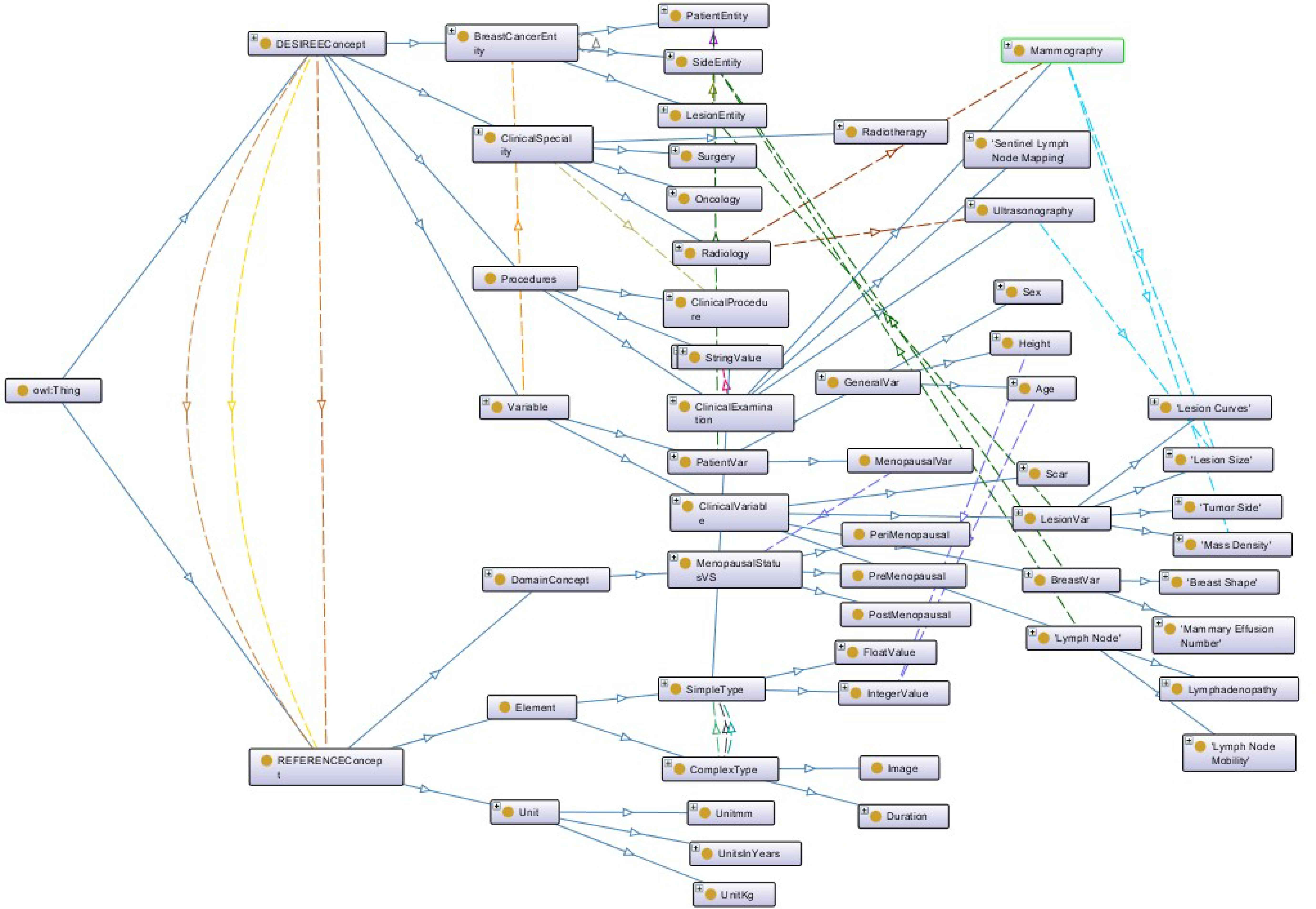
Semantic representation of breast cancer knowledge model
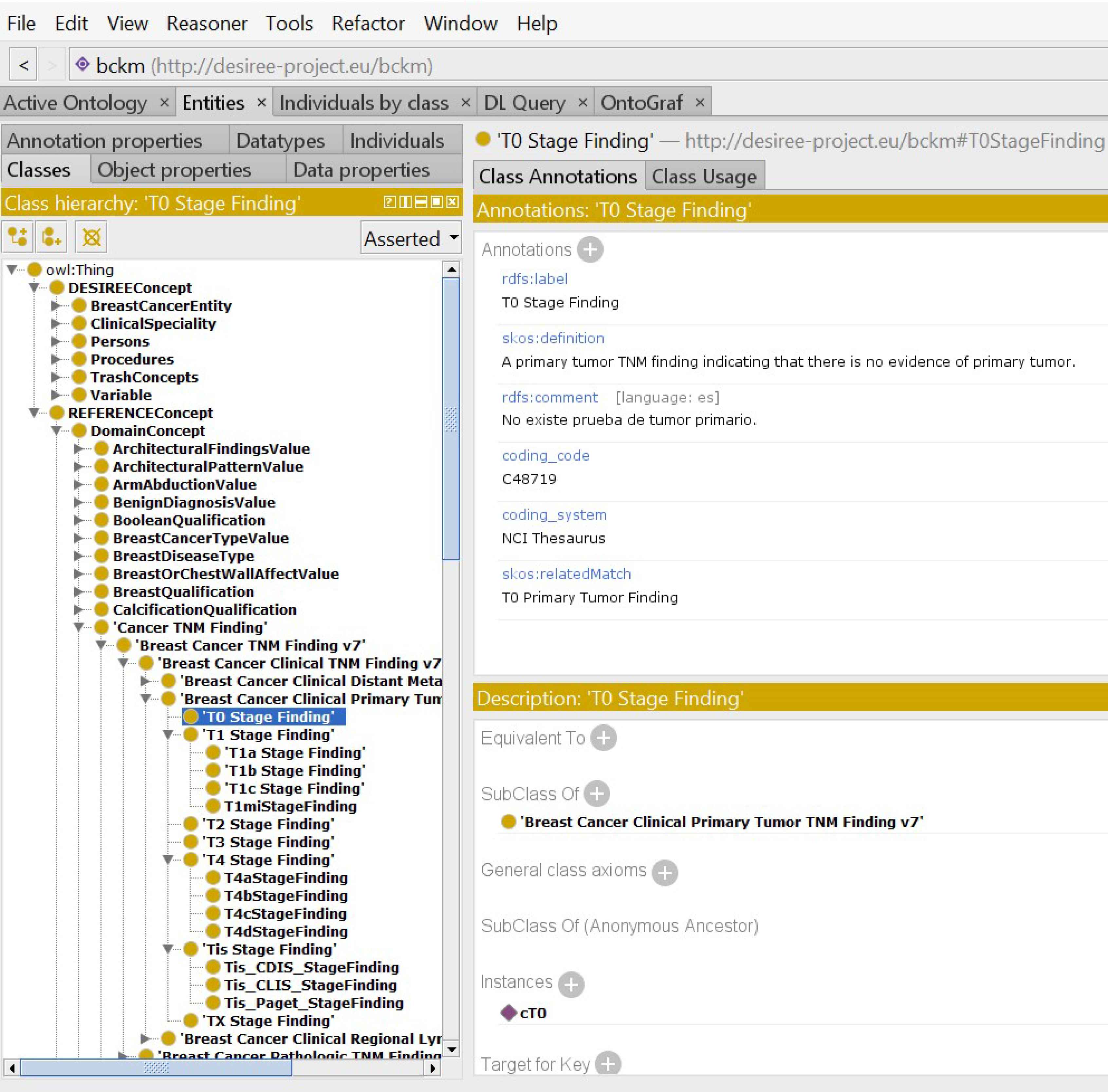
Example of a concept in the breast cancer knowledge model
As an overall architecture, the BCKM contains two main hierarchies, namely the DESIREE concept and REFERENCE concept. The DESIREE concept contains the entire specifications relevant to the DESIREE environment, namely the concepts relevant to the BUs. The REFERENCE concept contains the potential values for the terminologies under DESIREE concept. The DESIREE concept contains all the clinical data including the relevant clinical procedures, possible examinations, clinical findings, observations, etc. It also characterises the series of parameters and attributes under DESIREE concept into three main entities, namely Patient, Side, and Lesion entities, and allows the description of breast cancer patient cases. For example, this allows one to identify in a patient case whether the tumour is located on the left or right side of the breast, if a particular attribute is relevant to the patient (e.g. age, gender) or to the lesion (e.g. TNM staging, tumour size).
2.3. Similarity Analyser and Retrieval Algorithm
A high-level description of a generic CBR system is composed of four consecutive processes, namely retrieval, reuse, revise, and retain.8 With a collection of precedent cases and domain knowledge, one may implement one or more of these four steps in the application.
- •
Retrieval executes the retrieval algorithm with various similarity matrices to retrieve the case(s) that are most similar to the query case from the precedent ones present in the case base.
- •
Reuse or adaptation uses the knowledge and information acquired from the retrieved case(s) to solve the query case.
- •
Revise, through external means, revises the proposed solution.
- •
Retain learns from the problem-solving experience and stores in the case base the new information/knowledge acquired from the resolution of the query case for solving future new problems.
Among the four steps, as retrieval serves as the basis for the subsequent steps, implantation of an accurate case retrieval algorithm becomes crucial. In the present work, with jColibri software tool, the retrieval algorithm is defined using “local” and “global” similarity functions. Local similarity functions measure the distance between the simple attributes, whereas the global similarity function applies the results from local similarity measures to compare the compound attributes. In the proposed CB-DSS, a patient case is represented as compound attribute, composed of several simple attributes, including physiological and clinical attributes, such as age, gender, BIRADS, histological type, HER2 receptor, tumour size, etc. Thus, local similarity functions are first applied to compute the distance between simple attributes in the query case against the ones characterizing patient cases in the case base. The result of local similarity measures of all simple attributes are then aggregated using the global similarity function to select the patient case(s) that are most similar to the query case from the precedent ones present in the patient case base.
In the case retrieval algorithm, in addition to the simple similarity functions like numeric interval, equal, enumerated distance applicable to compare the simple attributes of the datatype, integer, string and enumerated types, the semantic similarity function is defined to avail the benefit of hierarchical placement of concepts in the domain ontology. Semantic similarity function computes the distance between the simple attributes of a query patient with those of patients retrieved from the patient case base as a degree of taxonomical proximity. Based on the taxonomical structure and location of concepts in the ontology, jColibri presents four semantic similarity functions,31 which are examined in the proposed CB-DSS. As shown in Eqs. 1 and 2, fdeep_basic and fdeep similarity functions take into account of the depth of the concept in the taxonomical structure of the ontology.
Where, LCS(i1,i2) is the set of the least common subsumer concepts of the two individuals, prof(Ci) is the depth of the concept Ci, and prof(i) is the depth of the individual and CN is the set of all the concepts in the knowledge model.
Likewise, Eqs. (3) and (4), cosine and detail similarity functions, measure the similarity between two vectors or sets, thus takes into account of the number of superclasses or ancestors in the ontology.
Where super(c,C) is the subset of concepts in C which are super concepts of c and t(i) is the set of concepts the individual i is an instance of.
Now, with a hierarchy tree of a DESIREE concept: “BreastTNMv7CT” from BCKM presented in Fig. 4, we describe how some of the key terms used in the above equations are computed.
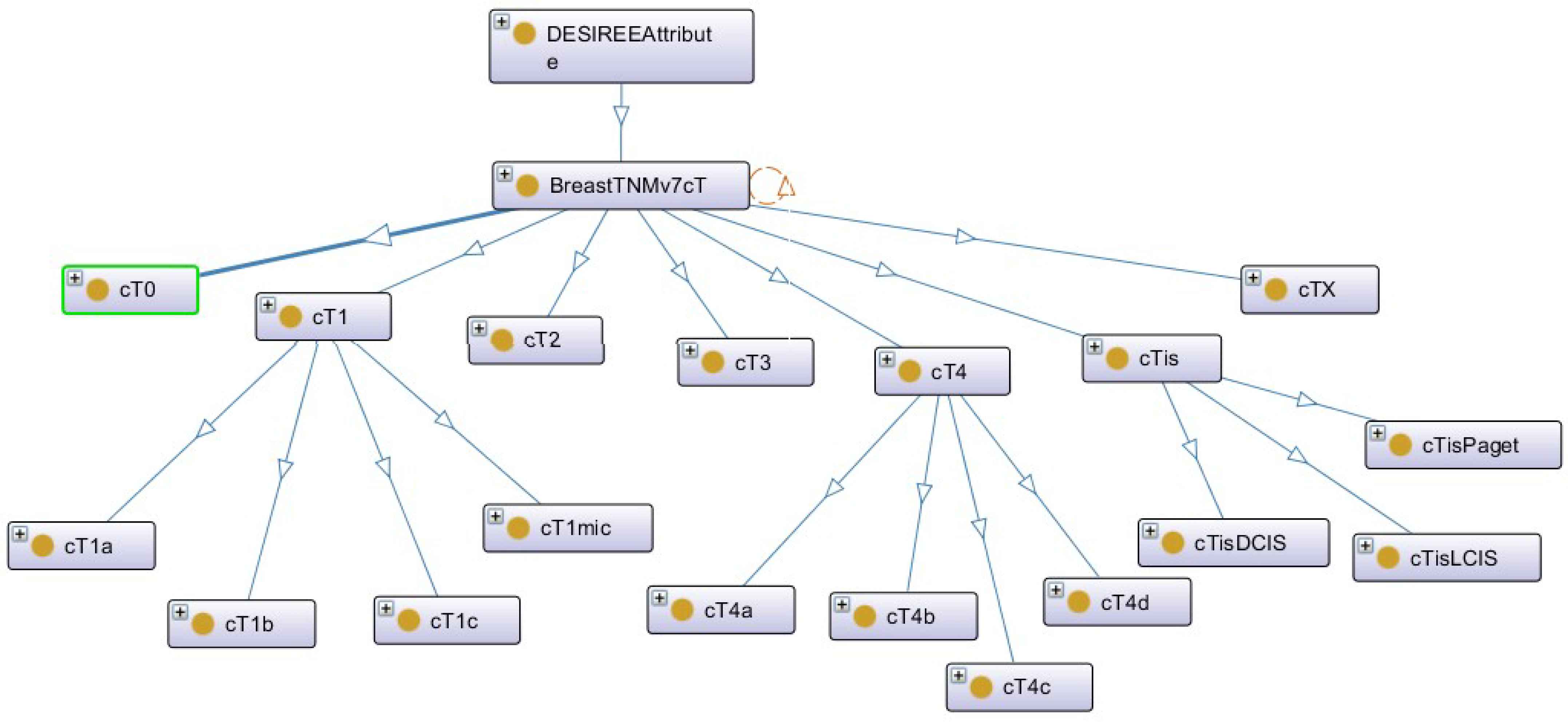
Hierarchy tree of the DESIREE concept – BreastTNMv7cT
In Eqs. 1 and 2, LCS(i1,i2) measures the similarity distance based on the most specific taxonomical ancestor common to i1 and i2. The more common the subsumer is, the more similar the terms will be, which ranges from ‘1’ for identical concepts to ‘0’. In our specific example, assume i1 is “cT1a” from the query case and i2 could correspond to any value ranging between “cT0” to “cTx” in the case base. For example, the most specific taxonomical ancestor common to “cT1a” and “cT1b” will be “cT1”, whereas for “cT1a” and “cT2” will be “BreastTNMv7cT”. Thus, the LCS(cT1a, cT1b) will be greater than LCS(cT1a, cT2).
In Eqs. 3 and 4, super(c, C) measures the similarity distance by taking into the account of the amount of shared superconcepts or ancestors of the pair under comparison. Higher the amount of shared superconcepts, shorter the similarity distance between the two concepts. Thus, according to Fig. 4, super(cT1a, cT1b) will take into account of the three supperconcepts “cT1”, “BreastTNMv7cT” and “DESIREEAttribute”, whereas super(cT1, cT2) will only take two superconcepts “BreastTNMv7cT” and “DESIREEAttribute” to compute the similarity distance. Therefore, super(cT1a, cT1b) will have a lower similarity distance than super(cT1, cT1).
Now, with local similarity measures computed for all the simple attributes using Eqs. 1 to 4, k-Nearest Neighbour (k-NN) is computed as the global similarity function to retrieve the top k similar cases from the patient case base. k-NN being a non-parametric lazy learning algorithm, it does not make any assumptions on the data distribution or require training data points to do any generalization. Given the query patient case xci and the local similarity measure computed for the patient cases in the case base yi, Euclidian distance between the query and case base is computed using Eq. 5.
Based on above computation, the k nearest patient cases are first located in the case base. The k-NN similarity measure is then computed using Eq. 6, which computes the arithmetic mean output across patient cases in the case base and returns a value between 0 ∼ 1, with 0 and 1 indicating the retrieved case being less and most similar to the query case.
2.4. Patient Case Representation
In a CB-DSS, the most important source of data is the set of resolved cases. Different applications may have different case representational requirements and as the size of the case base increases, it becomes critical that the CBR system accesses the stored cases efficiently. To address such challenges, jColibri provides persistence mechanism and in-memory organization for case base management.31
The persistence mechanism provides connectors that allow the CBR system to access the cases from the medium and return them in a systematic manner. The three different connectors include the case base connector, plain text connector, and ontology connector to manage the persistence of cases in the case base, textual files, and ontologies respectively. Secondly, inmemory mechanism deals with the data structure used to organize the cases in memory. To accommodate different data structure, including linear, tree structure, case retrieval nets etc., different CBR case base interfaces, including lineal case base, cached lineal case base, and ID indexed lineal case base are used.
In the present work, patient cases, which are currently simulated data using information from clinical partners as reference are stored using MySQL in a linear structure using lineal case base and the case base connector is implemented to access and retrieve cases from the case base. Hibernate library is internally used to execute the case base connector. To access data from the BCKM for semantic similarity measure computation, ontology connector is used. Internally, OntoBridge library is used to connect to the ontology.
The query patient case is stored outside the CB-DSS, within the DESIMS component. The exchange of patient data between the DESIMS and the CB-DSS is done through the HAPI FHIR server. Using the standard FHIR resources, including Patient, Observation, Condition, BodySite, and Specimen, the FHIR server retrieves and stores the query case from the DESIMS as a patient bundle. The patient bundle is then parsed and decoded by the CB-DSS for further analysis.
The attributes present in the patient case are divided into two case components, namely description component (patient description) and solution component (decision made by the BU). Similarity analysis is performed on the description variables to retrieve similar patient case(s) with description and solution variables. For example, in a patient case, the description component would include variables such as patient’s age, TNM staging, BIRADS, tumour size etc., whereas the solution component would include variables such as surgery, endocrine therapy, chemotherapy, and radiotherapy. In the present work, the patient cases present in the case base would include both description and solution components, whereas the query case would include only the description component.
3. A Case Study
To demonstrate the complete operation of the CB-DSS, we present a case study by testing a labeled query case. We consider a woman, aged 50, one pregnancy, one child, breast size = 95A, in premenopausal status. Clinical examination of left breast indicates one nodule of 25 mm at the union of external quadrants. Clinical exam of the ipsilateral axillary area shows one clinical node of size 10mm. Mammography shows the breast tumour size is 30 mm, BIRADS=4. Ultrasound exam shows one breast nodule of dimension 30 mm and one enlarged axillary lymph node of size larger than 10 mm. Microbiopsy of breast nodule indicates ductal invasive carcinoma, Estrogen receptor (ER)= 95%, Progesterone receptor (PR)= 60%, HER2= 0, SBR3, Ki67= 20% and tumoral cells are found in the cytopuncture of the axillary node. TNM staging is cT2, N1, and M0.
During run time, the query case is sent from the DESIMS to the CB-DSS through the FHIR server. The patient bundle from FHIR server is then parsed and decoded for further analysis by the CB-DSS. Attributes are read as the description component of the case. Depending on the data type of these individual attributes, different similarity functions are employed to measure local similarities. For example, for attributes with numeric data type, namely age, tumour size, ER, PR and Ki67, numeric local similarity functions are used. For attributes, which have a straight list of values namely gender, menopausal status, BIRADS category, etc., enumerated similarity function is used. Utilizing the knowledge codified in the BCKM presented in this paper, semantic similarity functions defined in Eqs. (1) ∼ (4) are used to measure local similarities, taking into account of the degree of taxonomical proximity in the ontology. The semantic similarity function is applied to attributes, namely breast TNM staging – cT, cN and cM, and histological type. Finally, when all local similarity measures are computed, the global similarity measure is computed using k-NN (Eq. 6), to retrieve the top k similar cases from the case base.
Retrieved patient cases are then compared with the query case through a rainbow box visual analytics GUI and displayed to the clinician/user. The CB-DSS results for the above query case are shown in Fig. 5. In rainbow boxes, each column corresponds to a patient: the “query” column with a white header represents the query patient, and the other columns represent the similar patients retrieved by the CB-DSS. The colour of the column headers indicates the type of treatment prescribed to these similar patients (here yellow for endocrine therapy and red for surgery). We can see that most similar patients were treated by surgery.
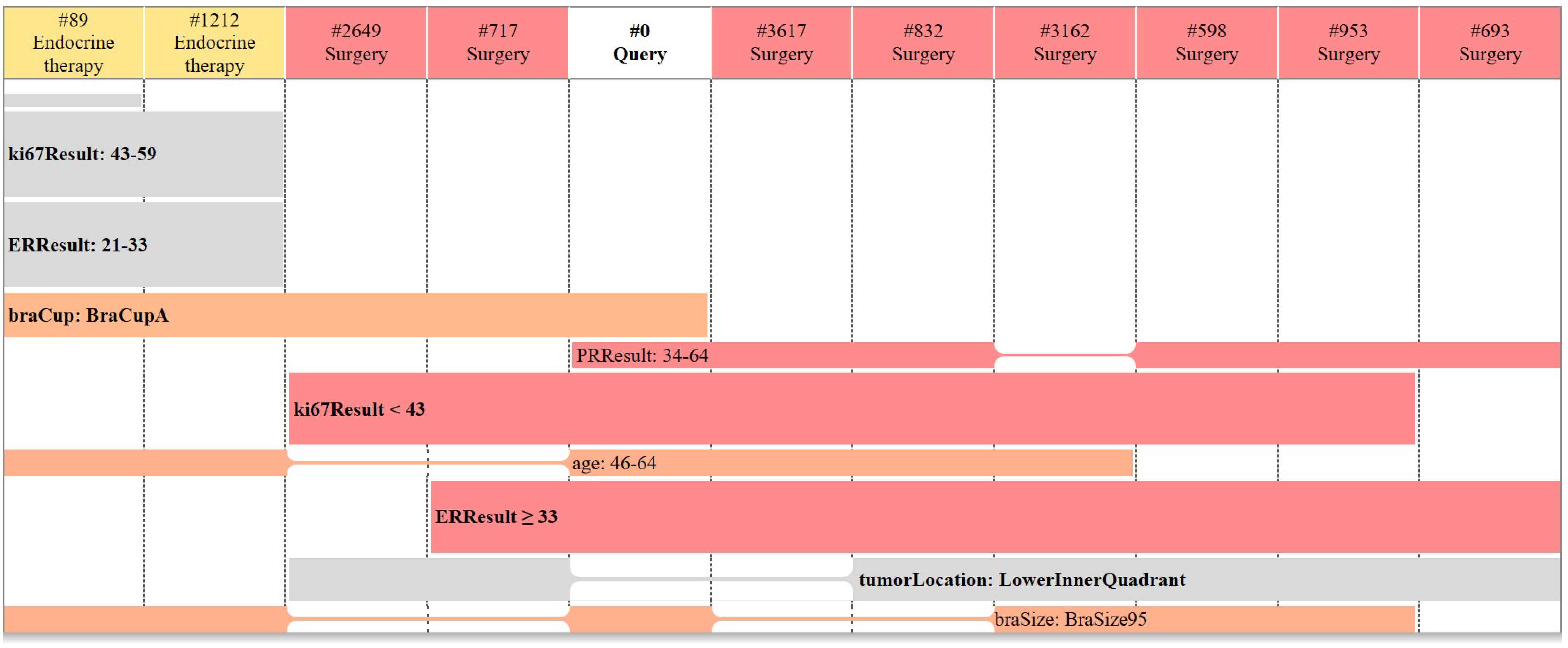
Visual analytics GUI for CB-DSS: results for the case study
The colored boxes below the column headers represent the characteristics shared by several patients. The boxes give the evidence of why patient cases are similar. A given box covers the columns corresponding to the patients sharing the characteristic: e.g. patients #89 and #1212 have a Ki67 value between 43 and 59. Holes appear in boxes when patients sharing a given characteristic are not contiguous (e.g. the PR result 34-64 box). Columns are placed in order to minimize the number of holes.
Boxes that do not cover the query patient are represented in grey color. The other boxes are colored and their color is the mean of the colors associated with the columns they cover. Thus, a box representing a characteristic present only in patients treated by surgery will be red (e.g., PR result 34-64), and a box mixing patients with surgery and endocrine therapy will be orange (e.g., age 46-64).
In Fig. 5, we can see that most similar patients were treated by surgery, and that most of the characteristics of the query patient (i.e. the colored boxes) are associated to red or reddish colors. Therefore, surgery seems the most appropriate type of treatment for the query case.
4. Conclusions
In this work, we have developed the CB-DSS module of the European-funded DESIREE project which aims at providing a web-based software for the therapeutic management of breast cancer. The proposed CB-DSS provides a tool for querying former cases in order to retrieve similar patient cases based on the defined case base and a case retrieval algorithm using a semantic similarity measure. The paper presents the overall framework of the proposed CB-DSS and systematically describes its workflow from clinical data integration to visualization of results to the user. We defined a BCKM to represent the domain knowledge as an ontology and we integrated a semantic similarity function to improve the similarity analyser and retrieval algorithm. To technically validate the proposed CB-DSS, the workflow of the framework is evaluated on a case study using a labelled query case. However, as the CB-DSS was developed based on simulated data generated using information from clinical partners, the medical relevance of the similarity measure, and the quality of the k most similar cases retrieved could not yet be validated. Further work is required, in terms of assigning weights to the attributes for computing local similarity measures, examining different similarity functions, and testing the relevance of the retrieval algorithm in a larger and reliable case base to clinically validate the proposed model.
Acknowledgements
The DESIREE project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 690238.
Footnotes
References
Cite this article
TY - JOUR AU - Booma Devi Sekar AU - Jean-Baptiste Lamy AU - Nekane Larburu AU - Brigitte Séroussi AU - Gilles Guézennec AU - Jacques Bouaud AU - Naiara Muro AU - Hui Wang AU - Jun Liu PY - 2018 DA - 2018/11/01 TI - Case-Based Decision Support System for Breast Cancer Management JO - International Journal of Computational Intelligence Systems SP - 28 EP - 38 VL - 12 IS - 1 SN - 1875-6883 UR - https://doi.org/10.2991/ijcis.2018.25905180 DO - 10.2991/ijcis.2018.25905180 ID - Sekar2018 ER -
