MicroRNAs associated with the pathways involved in the pathogenesis of abdominal aortic aneurysms
- DOI
- 10.1016/j.artres.2012.09.005How to use a DOI?
- Keywords
- MicroRNA; miRNA; Aneurysm; Vascular smooth muscle cell; Extracellular matrix
- Abstract
Objectives: MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at a post-transcriptional level. Through binding to mRNA sequences, miRNAs have a wide variety of functions, and are key regulators in vascular disease. Although there are only 2 papers looking directly at the association between miRNA and abdominal aortic aneurysms (AAA), several studies have looked at miRNAs implicated in vascular smooth muscle cell (VSMC) proliferation, extracellular matrix (ECM) remodelling, and the known genes and genetic loci associated with AAA. This review aims to determine potential miRNAs associated with the pathways involved in abdominal aortic aneurysm (AAA) pathophysiology, to guide future focused research.
Methods and results: A systematic review of the published literature was performed, searching for articles detailing miRNA associations with AAA or processes associated with aneurysm formation. Eighteen miRNAs were identified to be associated with aneurysm formation, ten miRNAs were associated with VSMC physiology, and nine miRNAs were involved in regulation of the ECM. Seven miRNAs were replicated in more than 1 study (miR-19b, miR-21, miR-26a, miR-29b, miR-146a, miR-221, miR-222).
Conclusions: The association between miRNAs associated with known AAA genes, and those involved in VSMC/ECM pathophysiology highlight promising areas for further significantly powered human studies, which with miRNA level modulation, present a novel opportunity to determine pathways for AAA formation.
- Copyright
- © 2012 Association for Research into Arterial Structure and Physiology. Published by Elsevier B.V. All rights reserved.
- Open Access
- This is an open access article distributed under the CC BY-NC license.
What this article adds?
This study concisely reviews current literature on the association between microRNA and both vascular smooth muscle cells and the extracellular matrix, known components of aneurysm pathophysiology. It also reviews current literature on microRNAs and abdominal aortic aneurysms, and the known genes and genetic loci associated with AAA. This thorough evaluation of the interrelation between microRNA and current knowledge regarding AAA formation highlights several areas of interest for further research.
Introduction
Aneurysm formation is a complex multifactorial process involving destructive remodelling of connective tissue throughout the affected segment of aortic wall.1 This process involves local chronic inflammation,2 a decrease in the number of smooth muscle cells in the tunica media,3 and fragmentation of the extracellular matrix of the aorta.4 During the last few years considerable effort has been devoted to the elucidation of the molecular mechanisms and pathways of abdominal aortic aneurysms (AAA) with recent work focussing on the role of microRNAs (miRNA). Due to the multiple post-transcriptional effects of individual miRNAs, it is possible that a single miRNA may be responsible for AAA, however it is more likely that a range of miRNAs will be identified associated with specific aspects of aneurysm formation. By understanding the pathophysiology of aneurysm formation, specific drug treatments can be developed to halt aneurysmal growth or prevent rupture. The study of miRNAs and their modulation will add to our understanding of AAA formation, and may yield potential therapeutic targets.
miRNAs are short (19–26) nucleotide non-coding RNA sequences, which are transcribed from DNA, however they are not translated into proteins. They are involved in human gene expression by binding to messenger RNA (mRNA) and preventing gene expression by the inhibition of protein synthesis.5 miRNAs were first discovered in 1993 in the lin-4 gene in Caenorhabditis elegans, where a 22 nucleotide RNA sequence was found to inhibit protein synthesis.6 In humans, let-7 was the first miRNA identified, which is involved in the control of developmental timing.7 miRNA sequences appear to be highly conserved among plants, micro-organisms and animals, suggesting that they represent an important regulatory pathway.8 There are currently over 1500 human miRNAs in the microRNA database9 which are thought to target approximately 60% of human genes.10
The aim of this review was to determine the extent of current knowledge regarding the role of miRNAs and their targets in the pathogenesis of abdominal aortic aneurysms, and to identify areas for future research. Medline, Embase and Health and Psychosocial Instruments databases were searched, using Ovid Online (Version: OvidSP_UI03.04.02.112, Ovid Technologies, Inc.) in January 2012. Although there are only 3 papers looking directly at the association between miRNA and abdominal aortic aneurysms (AAA), several studies have looked at miRNAs implicated in vascular smooth muscle cell (VSMC) proliferation, extracellular matrix (ECM) remodelling, and the known genes and genetic loci associated with AAA. This review aims to determine potential miRNAs associated with the pathways involved in abdominal aortic aneurysm (AAA) pathophysiology, to guide future focused research.
MicroRNA processing
In order to determine the role of miRNAs in aneurysmal disease it is important to understand miRNA biology. Mature miRNA is formed through a three step process. Initially, miRNA genes are transcribed into primary miRNA (pri-miRNA). Secondly, the pri-miRNA is cleaved into precursor-miRNA (pre-miRNA), and transported into the cytoplasm. The pre-miRNA is then cleaved and unwound, forming mature miRNA.11 Mature miRNA is an imperfect double strand due to a 3′ 2 nucleotide overhang on each end. Argonaute proteins, the primary components of the RNA-induced silencing complex (RISC) bind to the single stranded region of the functional miRNA strand,12 whilst the complementary strand is usually released and degraded.13 Rarely the complementary strand is also functional, becoming its own miRNA affecting a different mRNA.14 The miRNA-RISC complex binds to the target mRNA and results in translational repression, decapping, deadenylation or degradation15 of the mRNA, thus altering mRNA expression. As the interaction between miRNA and mRNA occurs between the 5′-UTR of the miRNA to the 3′-UTR region of the mRNA, a single miRNA can regulate hundreds of genes,16 and a single gene can be regulated by several miRNAs (Fig. 1). For example, the phosphate and tensin homologue (PTEN) mRNA, which is a key regulator in vascular smooth muscle cell proliferation and apoptosis, has been shown to be down-regulated by miR-19b,17 miR-21,18 and miR-221/222.19
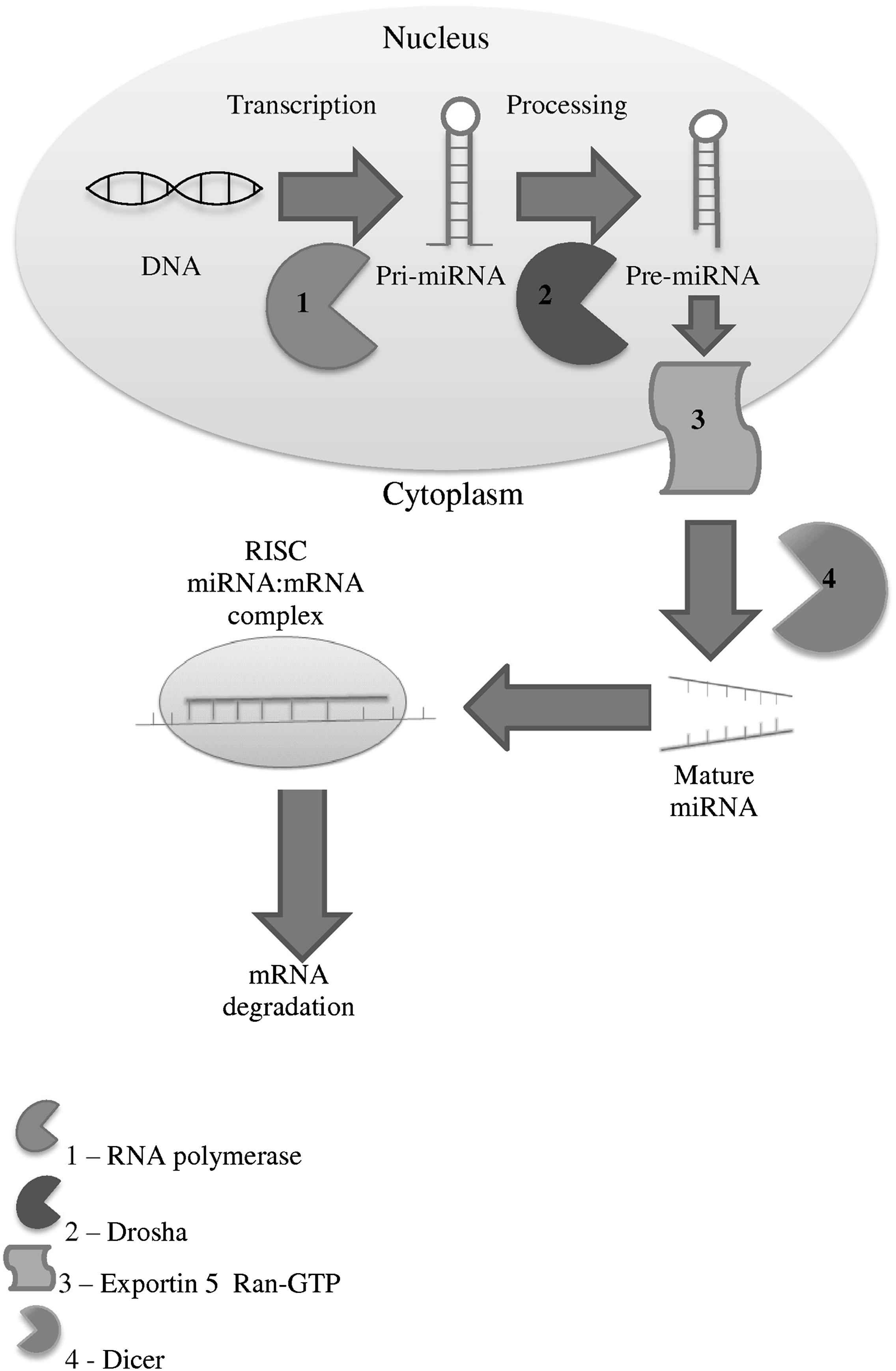
MicroRNA processing pathway.
The regulation of miRNA expression is not fully understood, with regulation thought to be at the transcriptional level. Studies have revealed that pre-miRNAs can be expressed throughout all tissue types; however mature miRNAs have a tissue specific expression, which suggests that there is a degree of post-transcriptional regulation.20 miRNAs are coded for in regions located within the introns of coding and non-coding genes,21 within exons22 and also in intergenic genomic repeats. The coding regions for many miRNAs have not been identified.23 miRNA coding regions may have their own promoters, and therefore are independently expressed, or they may occur in clusters (42% of cases), sharing the same transcriptional regulator.24
miRNAs associated with AAA
Maegdefessel et al.25 stated that decreased levels of miR-29b were associated with AAA formation. The paper studied the effects of the miR-29 family in two mouse models of AAA induction; porcine pancreatic elastase (PPE) infused mice and Angiotensin II infused mice. They determined that AAA induced mice had significantly decreased levels of miR-29b in aortic tissue samples, and went on to show that addition of anti-miR-29b in vivo increased collagen and elastin levels, but decreased MMP2 and MMP9 levels, whereas addition of pre-miR-29b normalised collagen and elastin levels, and increased MMP2 and MMP9 levels. This study also correlated their results with Human samples, comparing 15 Human AAA tissue specimens and 5 organ donor control samples. This revealed miR-29b to be significantly down-regulated in Human AAA, with these samples also containing significantly increased levels of collagen and elastin. This study contradicts previous work by Boon et al.,26 who demonstrated that vascular ageing and human thoracic aneurysms both have up-regulated miR-29 expression in the extracellular matrix, however this may be explained by the location of the aneurysmal aorta between these studies, or due to the study by Boon et al. looking at aortic tissue samples between young and aged mice. In vivo results from both Boon et al. and Maegdefessel et al. revealed that anti-miR-29 decreased the rate of aneurysm expansion in both studies, therefore it is possible that the increase in levels of miR-29 in the extracellular matrix leads to aneurysm formation, with intracellular levels of miR-29 being decreased as it is exported from the aortic smooth muscle cells. Maegdefessels study also contradicts the association between MMP 9 and AAA, identified in meta-analysis by Thompson et al.27 to be up-regulated in AAA, however this may solely be an effect of AAA induction in mice, as MMP levels were not assessed in Human aortic tissue.
An additional paper by Maegdefessel et al.28 identified that miR-21 expression increased as aneurysm size increased in murine AAA models. Over expression of miR-21 was shown to induce cell proliferation and decrease apoptosis in the aortic wall, decreasing the rate of aneurysm expansion, through decreasing the expression of PTEN, whereas the introduction of an antagomir targeting miR-21 lead to a marked increase in the aneurysm size. Maegdefessel also assessed the effect of nicotine on different miRNAs in AAA-induced mice, showing significant up-regulation of miR-21 and miR-26a, leading to down-regulation of PTEN in aortic tissue samples, with these results also identified in human aortic tissue samples.
Liu et al.29 identified fifteen differentially expressed miRNAs in a rat model of AAA, where aortic tissue was treated with either collagenase or saline, with a 50% increase in aortic diameter at 28 days considered an AAA. Initial microarray was carried out on 3 cases and 3 controls, identifying up-regulation of miR-146a, miR-221, miR-92a, miR-19b, miR-222, miR-34b, miR-142-3p, miR-188, miR-132, and miR-19a, and down-regulation of miR-301a, miR-152, miR-497, miR-148b-3p, and miR-181d. Four miRNAs (miR-19a, miR-19b, miR-132, and miR-222) were selected randomly for qRT-PCR validation using the same samples, confirming the microarray results. Although this study used small sample sizes, it determined significant results with stringent criteria, including a 2 fold change in expression, p < 0.001 and a false discovery rate <0.05.
The three studies above have all utilised different mouse models to induce AAAs therefore extrapolating their findings into definitive results which correlate with Human studies is not possible, however further investigation of these miRNAs in Human studies is warranted.
MicroRNAs associated with thoracic aortic aneurysms
Several papers have looked at miRNA expression in thoracic aneurysmal disease. In addition to Boon et al.26 identifying upregulation of miR-29b, a miRNome analysis on thoracic aortic dissection tissue by Liao et al.30 identified eighteen up-regulated and fifty-six down-regulated miRNAs, twelve of which were validated through PCR (upregulated miR-183, miR-433, miR-553, miR-491-3p, miR-30c, and miR-228-5p, down-regulated miR-143, miR-145, miR-22, miR-24, miR-93, and miR-768-5p). Sievers et al.31 identified up-regulation of miR-21, let-7g, and miR-145, and down-regulation of miR-214 and miR-125A-5p associated with aortic disease, using thoracic aortic aneurysm and acute aortic dissection tissue specimens. Jones et al.32 studied differential miRNA expression in human aortic tissue specimens from thoracic aortic aneurysms identifying five down-regulated miRNAs (miR-1, miR-21, miR-29a, miR-133a, miR-486-5p). MiR-143 and miR-145 were specifically studied by Elia et al.33 in thoracic aortic aneurysmal tissue, finding a significant down-regulation of both of these miRNAs in aneurysmal disease. Studies into thoracic aneurysms have therefore identified a total of eight upregulated and eleven down-regulated miRNAs, with miR-21 and miR-145 being both upregulated and down-regulated in different studies.
Although the underlying aetiology between abdominal and thoracic aortic aneurysms is not identical, both miR-21 and miR-29b have been found to be differentially expressed in both models, therefore further investigation into these miRNAs may provide additional information into aneurysm pathogenesis.
miRNAs regulating vascular smooth muscle cells
Vascular smooth muscle cells (VSMCs) play an important role in the pathogenesis of AAA. Increased VSMC remodelling occurs in AAA formation through a process of increased apoptosis in the medial layer of the aorta,34 and dedifferentiation of VSMCs,35 which permits increased VSMC proliferation,36,37 Several miRNAs have been identified which increase VSMC proliferation (up-regulation of miR-21, miR-221/222, miR-146a, miR-31, miR-26a, and miR-208, down-regulation of miR-1), and increase VSMC dedifferentiation (mir-143 and miR-145), therefore miRNA regulation of VSMCs is a plausible pathway leading to aneurysm formation.
Ji et al.38 undertook the first functional studies into the role of miRNAs in VSMCs, through experimental balloon angioplasty in rat carotid arteries, revealing significant up-regulation of miR-21. Down-regulation of miR-21 inhibited proliferation, thereby inhibiting neointima formation, however the exact mechanism is unknown. miR-21 has been extensively studied in cancer, being shown to both promote and inhibit cell proliferation in different tumours,39 and it has also been found to be essential for the differentiation of VSMCs through down-regulation of programmed cell death protein 4.40
miR-143 and miR-145 (co-transcribed miRNAs), which have been shown to be down-regulated in thoracic aortic aneurysms by Elia et al.,33 are involved in the regulation of VSMC phenotype41 with their knockout shown to alter the contractile phenotype of VSMC to a dedifferentiated, synthetic SMC phenotype that leads to neointima formation in aged mice.42 Chen et al.43 reported that myocardin overexpression in VSMCs induces the expression of miR-1, resulting in down-regulation of Pim-1 (a serine/threonine kinase), thus inhibiting VSMC proliferation.
Dedifferentiation and proliferation of VSMCs has also been found to be associated with miR-221, miR-222, and miR-146a.44 miR-221 exerts this effect through its interaction with platelet derived growth factor, causing VSMC proliferation, migration, and reduced expression of contractile genes.45 The expression of miR-146a has been found to be elevated in proliferative VSMCs, possibly through its interaction with Kruppel-like factor 4.46 These miRNAs have been found to be elevated in rat balloon injured arteries compared to uninjured controls,41 suggesting a role in VSMC proliferation and neointima formation.
Other miRNAs affecting VSMC pathophysiology include miR-26a, miR-31, and miR-208. miR-26a directly targets SMAD1 and SMAD4 (mothers against decapentaplegic homologue 1 and 4), thus promoting VSMC differentiation.47 miR-31 is an abundant miRNA in VSMCs, with its expression increased both in the proliferative state, and alongside neointimal growth.48 Furthermore, up-regulation of miR-31 has a pro-proliferative effect on VSMCs through the large tumour suppressor homologue 2 (LATS2), however miR-31 may be inhibited by mitogen-activated protein kinase (MAPK), suggesting the possibility of a MAPK/miR-31/LATS2 pathway.44 miR-208 increases basal and insulin-mediated VSMC proliferation through increasing the transformation of the cell cycle from G0/G1 phase to S phase.49
A total of 10 miRNAs have been identified in the above studies to be associated with VSMC pathophysiology. Despite these studies being conducted in non-human models and at anatomical locations distant from the infrarenal aorta where AAAs usually occur, this association between miRNAs and VSMC regulation is a potential target for future AAA research.
miRNAs in the extracellular matrix
Destruction of the extracellular matrix (ECM) is an established contributing factor for AAA formation, with evidence that the balance between matrix metalloproteinases (MMP) and their inhibitors is altered leading to matrix degradation.50 There are currently no studies determining miRNA involvement in the ECM in AAA disease specifically, however plausible biological candidates can be identified.
The miR-29 family (miR-29a, miR-29b, miR-29c) is a key regulator of the ECM. Evidence from the ECM of both trabecular meshwork cells51 and hepatic stellate cells52 have revealed that the miR-29 family causes down-regulation of fibrillins, elastin, collagens,52 and laminin53 with work on murine aortic development by Ott et al.54 replicating these results in the aortic ECM. miR-29b has also been found to directly target MMP2,55 which has been linked to AAA formation,56 along with the mRNAs for bone morphogenetic protein 1, disintegrin and metallopeptidase domain 12 and NF-Kappa-B inhibitor-interacting Ras-like protein 2.56 Results by Boon et al.26 demonstrated that vascular ageing and human thoracic aneurysms both have up-regulated miR-29 expression. As miR-29 has been shown to target Mcl-1 and p53, which induce apoptosis in cancer cells,57,58 and miR-29 is highly expressed in VSMCs59 as well as the ECM, Boon hypothesised that miR-29 may cause VSMC apoptosis, which is believed to promote aneurysm formation.60 In contradiction to this hypothesis, Maegdefessel et al.25 identified decreased levels of miR-29b in mouse models of AAA, and Human AAA tissue, however this was performed in aortic smooth muscle cells rather than the ECM, therefore miR-29 may cause VSMC apoptosis through interaction with the extracellular membrane, rather than through intracellular communications.
There is also evidence that miR-7 causes down-regulation of MMP2 and MMP9 in glioblastoma cell invasion,61 and miR-18a, miR-19a, and miR-19b decrease are associated with an increase in connective tissue growth factor (CTGF) and thrombospondin-1 with age-related heart failure, causing alteration of collagens type 1 and 3.62
Decreased levels of miR-133 and miR-30 have also been found to cause up-regulation of CTGF in heart disease and left ventricular hypertrophy, leading to collagen synthesis,63 showing their role in the control of structural change in the extracellular matrix of the myocardium.
Although there is no direct evidence that these miRNAs are differentially expressed in the ECM of aneurysmal tissue the 9 miRNAs above warrant further focused research into this specific area.
AAA associated miRNA prediction using genomic data
This review has identified 30 differentially expressed miRNAs involved either directly in AAA formation, or interacting with VSMCs or the ECM, with 7 miRNAs replicated in more than one study (miR-19b, miR-21, miR-26a, miR-29b, miR-146a, miR-221, miR-222). These results can be compared with validated genetic loci and genes known to be associated with AAA in order to extrapolate potential pathways in AAA formation, thus providing further insight into AAA pathophysiology and potentially enabling therapeutic intervention.
| Paper | Species | Case tissue | Control tissue | Cases vs control | Design | Up-regulated miRNAs | Down-regulated miRNAs |
|---|---|---|---|---|---|---|---|
| Liu et al., 2010 | Rat | Collagenase treated abdominal aortic | Abdominal aortic untreated control | 3 vs 3 | Micro-array and qRT-PCR for 4 miRNAs | 19a, 19b, 34b, 92a, 132, 142-3p, 146a, 188, 221, 222 | 148b-3p, 152, 181d, 301a, 497 |
| Maegdefessel et al. 2012 | Mouse and Human | AngII and PPE induced AAA in mice. Human AAA tissue |
Abdominal aortic untreated mouse control Human organ donor |
Human 15 cases vs 5 controls | qRT-PCR | 29b | |
| Maegdefessel et al., 2012 | Mouse and Human | AngII and PPE induced AAA in mice.Human AAA tissue | Abdominal aortic control | Not stated | Micro-array then qRT-PCR for specific miRNAs | 21 |
Results of literature review summarising all papers and abstracts associated with microRNA and aneurysms, with all differentially expressed microRNAs included, and those validated by qRT-PCR in bold.
There are currently three independently replicated genetic loci known to be associated with AAA formation (9p21,64 DAB2IP65 and LRP166), and three candidate genes for AAA that demonstrate significant association in meta-analyses (ACE, MTHFR, and MMP9).27 Little is currently known about the effects of miRNAs on these loci, genes and their transcripts, however several interactions have been identified in previous studies (Table 2) therefore it may be plausible to relate these to aneurysmal disease. From Table 2 it can be seen that ACE, MMP9, and LRP1 all have validated interactions with miRNAs identified to be potentially involved in aneurysmal disease, therefore analysis of the direct relationship between these genes and genetic loci and their associated miRNAs may yield significant results in determining AAA pathways.
| Locus/gene | Associated miRNAs | miRNAs associated with both locus/gene and aneurysmal disease |
|---|---|---|
| LRP1 | Let-7b, 1, 16, 30a, 30b, 30c, 30d, 30e, 155, 205, 338-5p, 545 | 1, 30 |
| DAB2IP | 338-3p, 338-5p | |
| 9p21 | ||
| MMP9 | 1, 21, 17, 17*, 26a, 29b, 30d, 92a, 132, 132*, 143, 143*, 145, 145*, 181a, 181a*, 181a-2*, 181b, 181c, 181c*, 181d, 188-3p, 188-5p, 206, 212, 218, 218-1*, 224, 376b, 451 | 1, 21, 132, 26a, 29b, 30, 92a, 143, 145, 181d, 188 |
| MTHFR | ||
| ACE | 16, 22, 22*, 29c, 29c*, 128, 132, 132*, 138, 195, 195*, 218, 222, 222*, 429 | 29c, 132, 222 |
Validated microRNAs targeting the associated genetic loci and genes found to be implicated in AAA formation in independently validated studies and meta-analyses. The results were identified by combining the searches for three validated miRNA databases, miRWalk,67 MiRTarBase,68 and miRecords,69 which map genes to microRNAs through published literature on PubMed. These results have been correlated with the 30 miRNAs identified in Table 1 as potentially dysregulated in aneurysmal disease.
miRNA target software is currently in its infancy, with validation databases requiring constant updating, and relying on published experimental data, therefore over time it is likely that more miRNAs will be identified associated with the above genetic loci and genes. An alternative method is using prediction software which determines potential interactions with miRNA, however these tools remain inaccurate with variation between databases, and more worryingly between prediction and validation results (i.e. some published interactions were not predicted). Although the authors feel that prediction methodology is likely to become useful to guide focused future research, it currently remains underdeveloped for this purpose, hence utilising literature from published material only in this review.
Conclusion
Although there remains a paucity of data looking directly at the association between miRNA and AAA, studies into the pathways known to be involved in aneurysm formation have yielded significant results which warrant further disease specific Human investigation. Thirty miRNAs were found to be potentially associated with AAA in this review, however there remains few consistent findings, with only 7 miRNAs validated in more than one independent study (miR-19b, miR-21, miR-26a, miR-29b, miR-146a, miR-221, miR-222). The correlation between these miRNAs implicated in aneurysmal disease, and those with VSMC pathophysiology, and ECM regulation highlight promising areas for future research.
Further detailed work is required in order to comprehensively assess and validate the association of miRNAs and AAA, as well as correlate the findings to the genes known to be involved in AAA formation. Although miRNome studies have been performed in murine models, larger Human miRNome studies must be carried out on both tissue, blood and plasma, to enable the translation of findings into blood based biological markers. These results can then be taken forward into cell culture and animal models so that specific therapeutic targets can be developed to stabilise the arterial wall in small aneurysms during the latent phase of aneurysmal growth, prior to definitive surgical correction.
miRNA level modulation represents an opportunity in the development of innovative treatments in AAA. The modification of miRNA levels in cell culture and animal models is currently promising, however much work is still required before these techniques may be applied in vivo.
Disclosures
None.
Funding
MJB is funded by a HEFCE Clinical Senior Lecturer Fellowship. JBW is funded by a fellowship from Heart Research UK.
Conflicts of interest
Nil.
References
Cite this article
TY - JOUR AU - P.W. Stather AU - J.B. Wild AU - N. Sylvius AU - E. Choke AU - R.D. Sayers AU - M.J. Bown PY - 2012 DA - 2012/10/11 TI - MicroRNAs associated with the pathways involved in the pathogenesis of abdominal aortic aneurysms JO - Artery Research SP - 28 EP - 35 VL - 7 IS - 1 SN - 1876-4401 UR - https://doi.org/10.1016/j.artres.2012.09.005 DO - 10.1016/j.artres.2012.09.005 ID - Stather2012 ER -
