Pyramidal Nonlocal Network for Histopathological Image of Breast Lymph Node Segmentation
- DOI
- 10.2991/ijcis.d.201030.001How to use a DOI?
- Keywords
- Deep learning; Histopathological image segmentation; Nonlocal network; Machine learning
- Abstract
The convolutional neural networks (CNNs) are frequently used in the segmentation of histopathological whole slide image- (WSI) acquired breast lymph nodes. The first layers in deep network architectures generally encode the geometric and color properties of objects in the training set, while the last layers encode the distinctive and detailed properties between classes. Modern segmentation approaches (DeepLabV3+, SegNet, PSPNet) are realized by evaluating these layers together. However, having a high parameter space of all these networks increases the calculation costs and prevents the researchers from working more effectively. In this study, we present a new pyramid-structured segmentation network (NonLocalSeg). Although the proposed network has low parameter space, its segmentation performance is similar to current architectures. The integration of the Nonlocal Module (NLM-a form of attention mechanism) or Asymmetric Pyramid Nonlocal block (APNB) into classical pyramid-built architectures has led to the reduction of network depth and narrowing of the parameter space while enabling coding of low and high image features. These mechanisms suppressed the unfocused background image, emphasizing the focused foreground object. As a result of a series of ablation experiments carried out, it is seen that the NLM and APNL mechanisms give the succeeded results. Although the network architectures adapting these mechanisms contain fewer parameters than current networks, it is observed that they have a similar accuracy (mean intersection over union [IoU]) range.
- Copyright
- © 2021 The Authors. Published by Atlantis Press B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
The digital pathology image is the digitization of slides which are tissues taken from the human body created by special processes using special scanners. These images are also called histopathological whole slide image (WSI). Pathological diagnosis is the examination of these images to determine whether there are diseases such as cancer. For the correct diagnosis of pathology, an accurate and reliable separation (segmentation) of tissues as normal or cancerous is essential.
With the proliferation of WSI data, many researches have worked on disease detection using machine learning methods [1–4]. As WSIs are large images, it is not possible to give a WSI directly into the machine learning model, considering GPU memory consumption. Commonly, when WSI is examined, small-sized images are analyzed independently of the whole. This method is called patch-based evaluation [5–7]. The image in Figure 1 shows patches from tumorous and normal tissues. Global content knowledge is lost in approaches that use only local patches, which hinders the detection of tumorous tissues.

Tumor and normal tissue sample patches in a whole slide image (WSI).
Through segmentation, pixels with similar color and texture properties in the image are grouped. This process, which is assigned an independent label value to each group, is used in medical images such as ultrasound, histopathological images, and so on. In recent years, convolutional neural networks (CNNs) have been used in segmentation studies and many special architectures have been developed [8–11] reports that the use of deep learning algorithms, especially CNNs, in breast histopathological image analysis is quite common. Although there are intensive studies regarding segmentation in histopathological images, the success of tumorous tissue segmentation in medical images is still not at the desired level. This can be explained by a number of factors. Firstly, the variety of tissues in medical images is extremely high. Figure 2 shows the tumor and normal tissue patches obtained in the histopathological WSI of breast lymph nodes. The color and texture differences are observed even among the images belonging to the same class. Secondly, the diversity of devices used in tissue preparation, lighting, and scanning increase the tissue variety. Even external factors such as dyeing and folding can also distort image quality. Figure 2 shows the texture folding (overlap) and stains. The mentioned factors make the tumorous tissue segmentation process difficult. As a result of these factors, an improved segmentation architecture is needed, which will not only explain the correlation between patches in the same class but also distinguish the differences between textures.
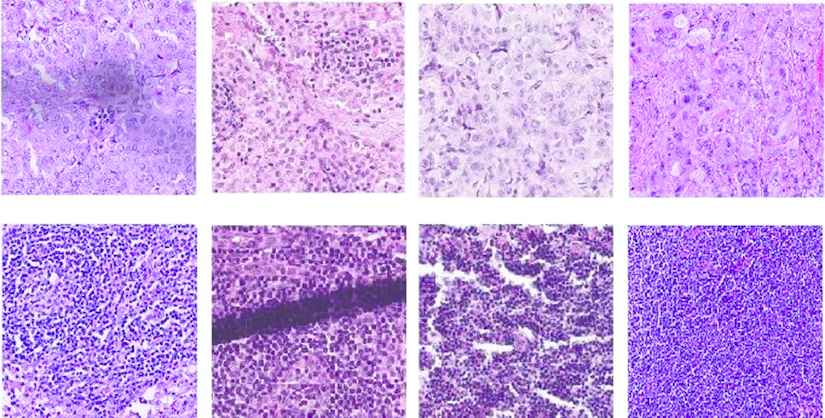
Collected sample tissues patches. Top: tumor, bottom: normal.
In pathological images, two different deep learning architectures are used to determine long-range dependencies between objects with different tissue structures and geometric shapes—stroma, cells, epithelium, tumor area. The first is deep convolutional networks with small kernel sizes; the second is shallow networks with large kernel sizes. In both convolutional networks, the number of hyperparameters and computational complexity are high.
A module called Global Convolution Network (GCN) can improve the aforementioned flaws in shallow networks with large kernel sizes. Using a more efficient method and without increasing computational costs, this module with large kernel sizes captures global contexts.
In addition, the use of attention mechanism is envisaged to increase the segmentation capability. It appears that the nonlocal network (NLN) module is used to accomplish this [12–14]. Wang et al. have developed the nonlocal module (NLM) combining CNN with traditional nonlocal means, which is used to reduce noise in the image. This module has been used to model spatial-temporal dependencies in video sequences and has increased the performance of existing networks. It is also used in the literature for image generation and segmentation problems [13].
Zhu et al. have demonstrated that the NLM has high GPU consumption and computing cost. They have developed the Asymmetric Pyramid Nonlocal Block (APNB). This block has been used in segmentation and provided high performance in terms of speed [14].
The innovative contributions of the proposed method can be summarized as follows:
A hybrid tumor segmentation architecture can capture global context information at a high speed. The architecture combines two important modules and an effective approach: GCN, NLM, and multi-scale segmentation approach.
Compared with state-of-the-art segmentation networks, the proposed network has reached a high mean intersection over union (IoU) value (68.6%) and has obtained a close pixel accuracy (PA) value. These results present the success of the proposed method for segmenting tumors in histopathological images of the lymph nodes of the breast.
The remainder of this paper is organized as follows: Section 2 provides a brief review of the related literature. We introduce the proposed hybrid architecture in Section 3. Section 4 lays out the experimental results and analysis. Discussion and future directions are given in Section 5. Finally, Section 6 presents a brief summary and our conclusions.
2. RELATED WORKS
2.1. Histopathological Image Segmentation
Fully convolution network (FCN) is the first image segmentation network using the convolutional layers. FCN has been developed by replacing the fully connected layers of standard CNNs used for classification with convolution layers [15]. This architecture has contributed to the development of many segmentation networks. Unet is one of the extended versions of FCN used in the medical images [16]. Layers close to the exit of the network makes the low-dimensional feature maps the same size as the input image using the deconvolution method. The segmentation accuracy is increased with skip connections. Skip connection is the addition of feature maps taken from different levels in the encoder to the decoder. Segmentation networks have been formed based on these networks [17–22].
There are some works segmenting tumors from histopathological medical images with interesting approaches. The work of Takahama et al. [23] is one of them. Patches obtained from WSI were given to the standard classification CNN to provide classification vector results. Then, these vectors were combined while taking into account the patch locations and given as an input to the segmentation network [9]. In another work is that the patches of different levels of WSIs were used for segmentation [10]. [24–26] are some of the current studies using CNN in the segmentation processes of histopathological image structures.
Hybrid network models in medical image segmentation have attracted a great deal of attention lately. For example, a segmentation architecture with fewer parameters has been developed by combining Unet, which is the classical and still the most popular architecture, with the residual learning framework for 3D MRI images segmentation [27]. In another study using a hybrid network, a stream that obtains shape information has been added to the classical Unet architecture [28].
2.2. Encoder–Decoder Architecture
The standard autoencoder architecture has an encoder and a decoder module. The encoder module is the part that gradually reduces feature maps size and captures advanced semantic information. The decoder module, on the other hand, is the module that gradually converts discriminative features (low-level resolution) learned in the encoder module to high-resolution. This architecture is used in many different areas of image processing such as object detection [29], pedestrian detection [30], facial landmark detection [31], and semantic segmentation [16,32,33]. In image segmentation, newly developed modules are added to the features taken from different scales in the encoder [34–37].
2.3. Effective Convolution Modules
Filter masks with high kernel dimensions are used to detect the relationships between objects in high resolution images. However, increasing kernel size causes the expansion of the parameter space, thereby increasing the cost. GCN is a way to effectively use large kernels [12,36,38]. The internal structure of the GCN module is shown in Figure 3(a). It is based on the parallel use of different-sized convolution layers. Considering the k kernel size, the cost of convolution with the kxk mask is quite high. Instead, the combination of
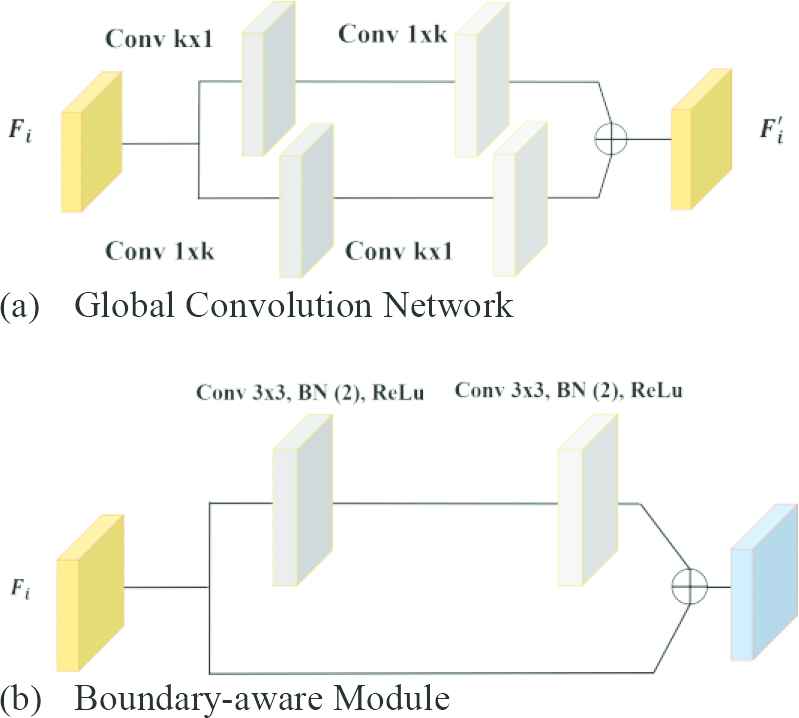
Effective convolution modules: (a) Global Convolution Network (GCN) module and (b) Boundary-aware Module (BM).
The convolution process causes shifts, scatter or thinning on the edge pixels (boundary regions) of the image objects. This situation causes problems in terms of perception. The boundary highlight module also called Boundary-aware Module (BM) is used to overcome such problems [12,36]. BM improves these disorders that occur in boundary information as a result of convolution. BM is integrated into the proposed network structure during the training. The internal structure of this module is given in Figure 3(b).
2.4. Nonlocal Module
NLM was first used in video classification. It was formulated with inspiration from nonlocal means used to reduce noise in the image. It concerns the self-attention (SA) module. SA mechanism allows inputs to interact with each other and find out which they should pay more attention to. SA was introduced as a module in language translation networks and later used in the field of images [39–42]. SA can be viewed as a form of the nonlocal mean [13].
3. METHOD
Potential differences can be found between the feature representations of the pixels of the same class. These differences cause inconsistency in the segmentation result when the contextual information of the image is not well coded [12]. NLM by analyzing the problem in this context is intended to create relationships between features.
The proposed network is based on fully convolutional network-based encoder–decoder architecture. The encoder achieves semantically effective features from the image and the decoder uses the achieved features to make semantic segmentation. The architecture of the network is given in Figure 4. The multi-stage segmentation strategy developed by Zhao et al. is used in the proposed network [36].
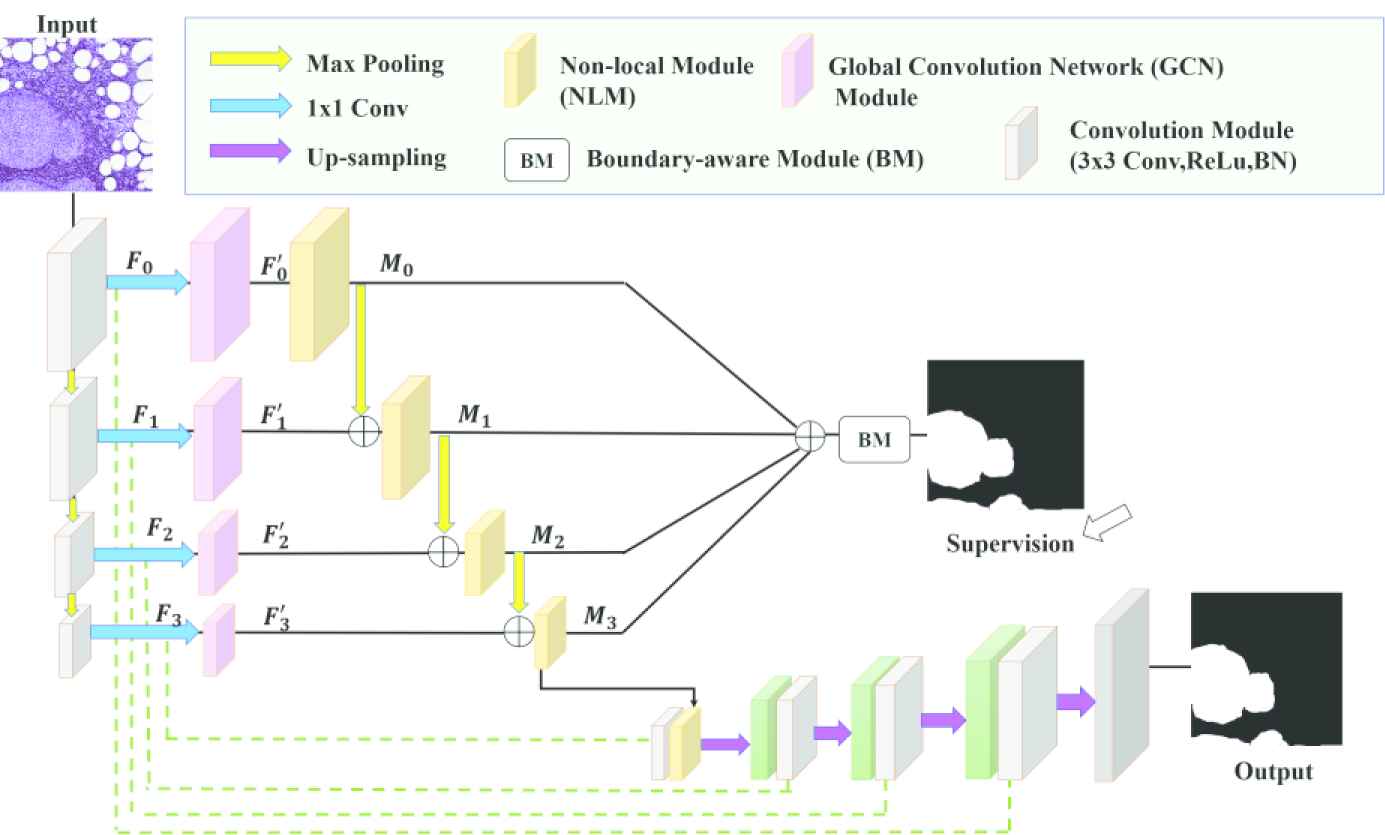
Pyramid nonlocal network architecture (NonLocalSeg).
Between the stages of the decoder, 3 × 3 kernel convolutions, ReLu, and batch normalization, and finally 2 × 2 max pooling have been performed. Table 1 the dimensions of input and output features of the stages are given.
| Stage | Input | Output |
|---|---|---|
| F0 | 3 × 2562 | 64 × 1282 |
| F1 | 64 × 1282 | 128 × 642 |
| F2 | 128 × 642 | 256 × 322 |
| F3 | 512 × 322 | 1024 × 162 |
Input and output dimensions of the stages.
In each stage, the feature maps channels are reduced through the 1 × 1 convolution layer. We call the feature maps {Fi} in the
3.1. Large Kernel Convolution
In the proposed network, different-sized kernels are used for the modules (GCN), where feature maps pass through each stage. The dimensions of the feature maps passing through the stages decrease as they progress. General contexts are captured using size-appropriate kernels for feature maps that are shrinking in size. The kernel sizes are 15, 7, 5, and 3 respectively for F0, F1, F2, and F3.
3.2. Nonlocal Module
Standard NLM is given Figure 5. [13]. Let denote
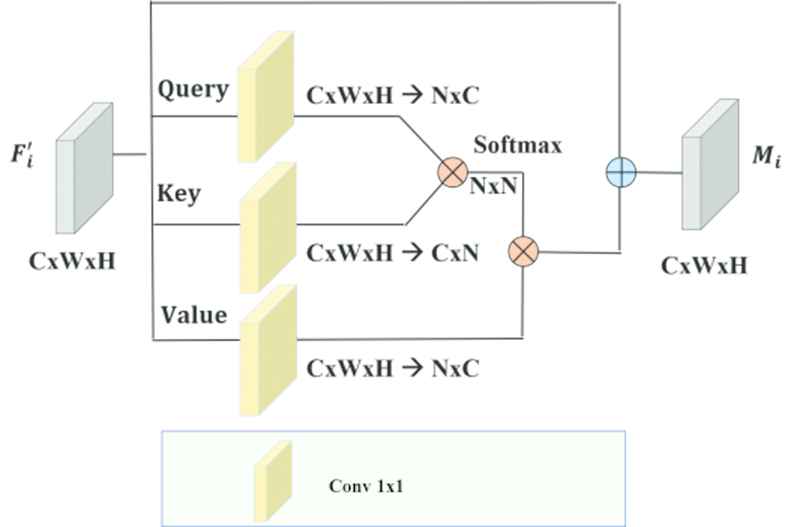
Standard nonlocal module (NLM).
Then the similarity matrix
Afterward, normalization is applied to V to get a unified similarity matrix as
As indicated by [13], the normalizing function f can take the structure SoftMax. SoftMax is comparable to the SA system and demonstrated to function admirably in numerous tasks, for example, machine translation and image generation.
For every location in γ, the output of the attention layer is
Feature maps
3.3. Asymmetrical Pyramid Nonlocal Block
It is observed that Eqs. (2) and (4) calculation costs are high for standard NLM. The two matrix products have
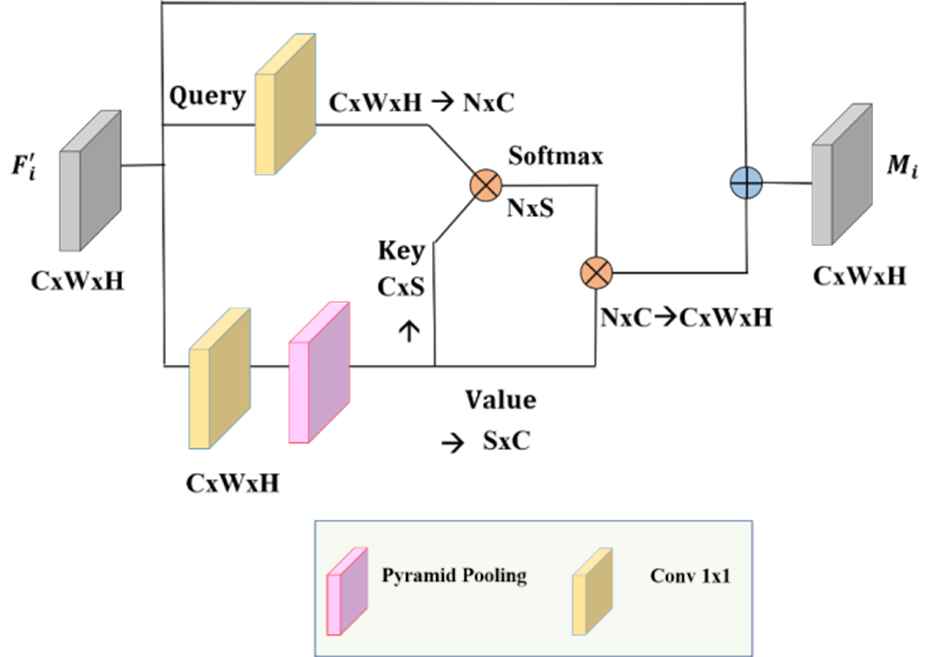
Asymetrical pyramid nonlocal block (APNB).
Returning to the design of the NLM, changing N to a small number S is equivalent to sampling several representative points from θ and γ instead of feeding all the spatial points, as illustrated in Figure 6. Consequently, computational complexity could be considerably decreased.
APNB is faster than standard NLM. The number of feature maps channels was reduced using Spatial Pyramid Pooling (SPP) as shown in Figure 6. SPP is used as a sampling module. In this study, NLM is replaced with APNB to analyze its impact on segmentation accuracy.
3.4. Pyramidal Feature Gathering for Segmentation
Once the encoder has acquired features that contain contextual information, the decoder must effectively combine these features to complete the segmentation. The merging process starts with the addition of low-dimensional feature maps. As seen in Figure 4, the features containing contextual information combine with features containing spatial information from the encoder. After increasing the size of feature maps by up-sampling, it combines with feature maps from the previous stage.
3.5. Deep Supervision
Another important factor affecting success in segmentation is to use the segmentation results obtained from different stages of the network during training [43]. This process is called supervision and we have applied this extra control in the proposed network. Overall, the process is used as an additional cost in training by combining the prediction results obtained from the network stages. Thus, the middle layers of the network learn to distinguish semantic discriminatory features. There are similar studies in the literature [8,36,44]. Considering all the losses, the total objective function is
3.6. Dataset
WSI used in this study was obtained from the contest named CAMELYON16 in 2016. WSI has 40× lens magnification. The average size of images is 1 GB and dimensions are 200,000 × 100,000. They are in the TIFF file format with different resolution levels.
The aim of the contest is the automatic tumor detection in the histopathological images of lymph nodes taken from the breast cancer patient. The areas with tumors in all WSIs were determined under the supervision of expert pathologists and recorded as two-level images (masks). The competition organization gave the position of tumor on slides to the researchers in XML file format. The slide images prepared in the pyramid structure have 8 levels between 0 and 7 in total. Mask images of WSIs can be obtained using the provided files. Table 2 provides detailed information about the dataset [45].
| Normal | Tumor | Total | |
|---|---|---|---|
| Train | 160 | 111 | 271 |
| Test | 80 | 49 | 129 |
| Total | 240 | 160 | 400 |
CAMELYON16 dataset.
From the Camelyon16 training set, a total of 8000 patches with tumor and normal tissues were collected from 111 Tumor WSIs. These patches created were used in the training of networks. From the Camelyon16 test set, a total of 2000 patches with tumor and normal tissue were collected from 25 Tumor WSIs. These patches created were used to obtain the test results of the networks. The size of all patches in the data set is 512 × 512. The total patches size for the used dataset is 10.000.
Random image cropping, one of the data augmentation techniques, was used in the training and testing stages of networks. As an input to the networks, randomly cropped images with 256 × 256 size from 512 × 512 images are given. Data augmentation techniques are used in the training [46].
Figure 7(a) shows a WSI structure, it has many levels as shown. In Figure 7(b) shows the WSI 3rd level's patch containing tumor and normal tissue. Finally, Figure 7(c) shows the mask of the image in (b). The white part represents the tumor. The size of the images in the figure (b) and (c) is 512 × 512.
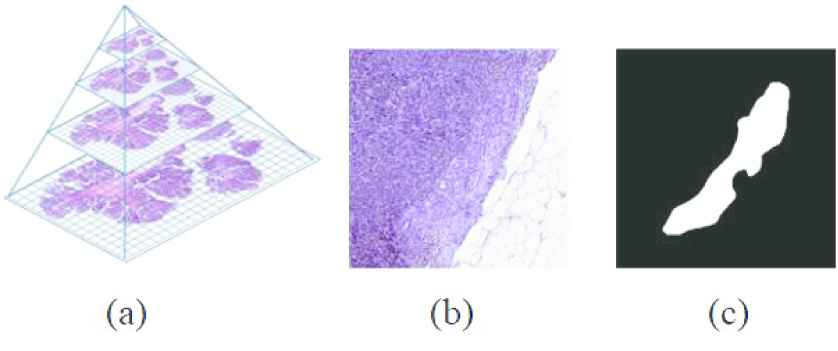
(a) The structure of a whole slide image (WSI), (b) a patch of level 3 of WSI, and (c) a mask of the patch.
4. EXPERIMENTS
4.1. Experimental Setting
In this section, we have provided dataset, training details and evaluation metric used in networks.
4.1.1. Training and implementation details
The proposed network is trained with Adam optimizer, and weight-decay = 0.0005. In the experiments, the weight layers are started without any pretrained parameters and the initial bias is set to 0. At the same time, all the networks in Table 3 are run 10 epochs. The learning rate initial value is 0.0001, after the 5th epoch, the learning rate is divided by 10. We adopt dropout after each fusion of the NLM and GCN [47]. We fine-tune our models with BatchNorm [48] enabled when it is applied (batch size is 2). Using the Pytorch library, we used a computer with a Nvidia Quadro M4000 (under CUDA 9.0 without other ongoing programs) graphics card with 8GB of GDDR5 GPU memory. Windows Server 2012 R operating system is installed on the computer.
| Methods | Backbone | Pretrained | Mean IoU (%) | PA (%) |
|---|---|---|---|---|
| DeepLabV3+ [32] | Xception | Yes | 61.9 | 85.6 |
| FCN8s [15] | VGG16 | Yes | 15.1 | 30.3 |
| UNet [16] | No | 58.5 | 79.1 | |
| PSPNET [49] | ResNet101 | Yes | 53.1 | 78.0 |
| ICNet [34]a | No | 49.2 | 66.4 | |
| SegNet [33]a | VGG19 | Yes | 61.6 | 81.9 |
| NonLocalSeg | No | 68.6 | 85.1 |
(a) Real-time segmentation networks.
Bold indicates the success of the method used.
Comparison with current segmentation networks.
4.1.2. Evaluation
IoU, also known as the Jaccard Index, is the standard measure of accuracy in semantic segmentation. IoU is measured as the ratio between the underlying truth and the predicted segmentation mask. The mean IoU is calculated by summing all the IoUs and dividing by the number of classes. In Eq. (11), let
Another measurement metric for segmentation is PA. The formula of the PA is given Eq. (13). PA is the percent of pixels in the image that are classified correctly.
Abbreviations our case TP denotes correctly classified pixels, FN denotes pixels not detected, and FP denotes background pixels classified as parts of a class. And finally, TN denotes the background pixels which are classified correctly.
4.2. Result
4.2.1. Ablation study on the proposed model
We have made some ablation studies on the proposed network and compared the results in terms of accuracy (mean IoU, PA) and speed (second). Ablations results are given in Table 4. The efficiency of the modules (NLM, APNB) integrated into the GCN module base network has been investigated. The network with NLM increases the performance of the base network from 62.2% to 68.6% (mean IoU). By replacing NLM with APNB a minor performance decrease (68.6% → 67.7%). Table 4 also shows the PA values of the networks. In experimental studies, it has been observed that the network with NLM has the highest PA value.
| With NLM | With APNB | Without NLM and APNB | |
|---|---|---|---|
| Mean IoU (%) | 68.6 | 67.7 | 62.2 |
| PA (%) | 85.1 | 84.1 | 82.5 |
| Avg. Execution Time (second) | 0.040 | 0.035 | 0.028 |
NLM, nonlocal module; APNB, asymmetric pyramid nonlocal block.
Ablation study on the proposed network.
In Table 4, the average execution time of the networks has been given in second for a 256 × 256 × 3 histopathological image. The network with NLM is the slowest compared to others, which is expected that because more products are made in NLM.
4.2.2. Comparison to state-of-the-art networks
A large number of epochs are needed to converge the training of networks with a high number of parameters. The proposed network has been converged faster than all the networks we have trained using the same epoch number. The success of the proposed network (68.6% mean IoU) has been shown in Table 3. Using low parameter space and the effective modules for a segmentation network have yielded better results. In Table 3, the 5th column gives the PA measurement values. The DeepLabV3+ network has the highest PA value. The proposed network has achieved the second-highest PA value, with a slight difference between the highest PA value.
Table 5 shows the number of parameters (million-M) of the successful networks that have passed the 60% mean IoU threshold and the average execution time spent to predict an image segmentation mask. The proposed network has the least number of parameters and is the fastest one. The average execution time of the proposed network is 0.040 seconds for a 256 × 256 × 3 histopathological image.
| Methods | Avg. Execution Time (Second) | # Param (M) |
|---|---|---|
| DeepLabV3+ [32] | 0.148 | 54 |
| SegNet [33] | 0.072 | 9 |
| NonLocalSeg | 0.040 | 6 |
Comparison of the execution time and the number of parameters.
Figure 8 in Section 7 shows the predicted segmentation masks of networks which have more than 60% of mean IoU. The first and second columns of Figure 8 show histopathological images and their ground truth masks. In the remaining columns of Figure 8, the tumor probability masks produced by Deeplabv3+, SegNet, and the proposed network are given respectively. When the tumor probability masks produced by the networks are examined in Figure 8, it is seen that they are different from the ground truth masks. However, it is determined that the mask results of the proposed network closer to the ground truth masks. The edges of the SegNet results are not clear. In DeepLabv3+, it is seen that tumor and normal tissue borders are intertwined.
5. DISCUSSION AND FUTURE RESEARCH
The segmentation process is difficult in histopathological images due to heterogeneous structure and tissue diversity. We propose a segmentation network model has been proposed model to make this process faster and more successful than existing networks. By keeping the parameter space of the network lower, the network has been trained faster. A low epoch value is used to train all networks. The results are presented in tables and figures. The comparison networks and the proposed network's visual results are given in Figure 8. When the visual results of all networks are compared with the ground truth masks, the one-to-one similarity is not obtained. However, the visual results of the proposed network are the most similar to ground truth masks.
Additionally, the shortest average execution time belongs to the proposed network. Speed is crucial for high-resolution images (WSI) analysis so that fast segmentation results can be obtained.
For future work, some tactics are used during training to improve the performance of networks in segmentation. For example, [35,37] use higher batch size and epoch value during training. The impact on network performance will be investigated using these tactics.
6. CONCLUSION
In this study, we implement a novel architecture of focus for the segmentation of histopathological images. This architecture network integrates a multi-scale strategy to combine semantic information at various levels and NLM to gradually combine relevant contextual features. To approve our strategy, we conduct the test on WSIs from the CAMELYON16 Challenge. We give extensive tests to examine the impact of the individual components of the proposed architecture. In addition, we compare our model to existing methods that have been lately present for the natural scene and medical image segmentation. The achievement of our method may be explained by having an enhanced ability to model rich contextual dependencies over local features of images.
The proposed segmentation network integrates GCN [12], NLM [13], APNB [14] modules and uses pyramid feature fusion [36] and segmentation supervision [24,37] strategies. Finally, the proposed network model shows that it is a valid tool to assist in rapid tumor segmentation, with its low parameter size and performance of segmentation close to current segmentation networks.
7. QUALITATIVE RESULT


Qualitative results of the networks are given. The first two columns show input images and ground truth masks. The remaining columns show probability masks of DeepLabv3+, SegNet, and NonLocalSeg networks, respectively.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHORS' CONTRIBUTIONS
Zehra Bozdag contributed to the design and implementation of the research, to the analysis of the results. Zehra Bozdag wrote the manuscript in consultation with Fatih Talu.
Funding Statement
The author(s) received no financial support for the research, authorship, and/or publication of this article.
REFERENCES
Cite this article
TY - JOUR AU - Zehra Bozdağ AU - Fatih M. Talu PY - 2020 DA - 2020/11/06 TI - Pyramidal Nonlocal Network for Histopathological Image of Breast Lymph Node Segmentation JO - International Journal of Computational Intelligence Systems SP - 122 EP - 131 VL - 14 IS - 1 SN - 1875-6883 UR - https://doi.org/10.2991/ijcis.d.201030.001 DO - 10.2991/ijcis.d.201030.001 ID - Bozdağ2020 ER -

