Baseline Photos and Confident Annotation Improve Automated Detection of Cutaneous Graft-Versus-Host Disease
 , Kelsey Parks1, 3,
, Kelsey Parks1, 3,  , Inga Saknite3,
, Inga Saknite3,  , Tahsin Reasat2,
, Tahsin Reasat2,  , Austin D. Cronin1, 3, Lee E. Wheless1, 3,
, Austin D. Cronin1, 3, Lee E. Wheless1, 3,  , Benoit M. Dawant2, Eric R. Tkaczyk1, 3, 4, *,
, Benoit M. Dawant2, Eric R. Tkaczyk1, 3, 4, *, 
- DOI
- 10.2991/chi.k.210704.001How to use a DOI?
- Keywords
- Cutaneous erythema; chronic graft-versus-host disease; change detection; longitudinal photos; baseline photos; computer vision; convolutional neural networks
- Abstract
Cutaneous erythema is used in diagnosis and response assessment of cutaneous chronic graft-versus-host disease (cGVHD). The development of objective erythema evaluation methods remains a challenge. We used a pre-trained neural network to segment cGVHD erythema by detecting changes relative to a patient’s registered baseline photo. We fixed this change detection algorithm on human annotations from a single photo pair, by using either a traditional approach or by marking definitely affected (“Do Not Miss”, DNM) and definitely unaffected skin (“Do Not Include”, DNI). The fixed algorithm was applied to each of the remaining 47 test photo pairs from six follow-up sessions of one patient. We used both the Dice index and the opinion of two board-certified dermatologists to evaluate the algorithm performance. The change detection algorithm correctly assigned 80% of the pixels, regardless of whether it was fixed on traditional (median accuracy: 0.77, interquartile range 0.62–0.87) or DNM/DNI segmentations (0.81, 0.65–0.89). When the algorithm was fixed on markings by different annotators, the DNM/DNI achieved more consistent outputs (median Dice indices: 0.94–0.96) than the traditional method (0.73–0.81). Compared to viewing only rash photos, the addition of baseline photos improved the reliability of dermatologists’ scoring. The inter-rater intraclass correlation coefficient increased from 0.19 (95% confidence interval lower bound: 0.06) to 0.51 (lower bound: 0.35). In conclusion, a change detection algorithm accurately assigned erythema in longitudinal photos of cGVHD. The reliability was significantly improved by exclusively using confident human segmentations to fix the algorithm. Baseline photos improved the agreement among two dermatologists in assessing algorithm performance.
- Copyright
- © 2021 International Academy for Clinical Hematology. Publishing services by Atlantis Press International B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Skin is the most commonly affected organ in chronic graft-versus-host disease (cGVHD), a leading cause of long-term non-relapse mortality and morbidity after allogeneic hematopoietic cell transplantation (HCT) [1,2]. Tracking change in cutaneous manifestations is critical to evaluate treatment efficacy or disease progression. Erythema is a common manifestation of cutaneous cGVHD assessed visually as the affected body surface area (BSA). Reversal of erythema has been associated with improved survival [3,4]. However, it is estimated that a clinical exam can only detect a minimal change in erythema of 19–22% BSA [5].
Computer-aided methods could provide accurate and objective assessment of skin change. However, their development for cGVHD is challenging due to the heterogeneity of cutaneous presentations and the lack of biological ground truth, as even normal-appearing skin can have microscopic GVHD features [6]. Traditionally, segmentation algorithms are developed from annotations by expert clinicians. The annotator marks the rash-affected area, and the remaining area is considered unaffected, regardless of how confident the annotator is in their marking. Each human annotator may have a different threshold for what they consider affected [7], resulting in high variability. This limits the development of computer-aided algorithms, which generally assume confident human inputs. Baseline photography is commonly used to detect change in neoplastic skin lesions (e.g. melanoma surveillance) [8,9]. However, its value is unknown in detecting non-neoplastic skin change.
In this study, we aimed to develop a neural network-based algorithm that provides a precise and objective measurement of change in cGVHD erythema. We hypothesized that limiting human annotation to areas of confidence in an image increases algorithm accuracy. We also hypothesized that viewing baseline photos improves reliability.
2. MATERIALS AND METHODS
2.1. Image Acquisition and Preparation
We acquired 56 pairs of 3D photographs from a hematopoietic cell transplant patient using a Canfield Vectra H1 camera. Through stereophotogrammetry technology, the camera captures a 165 × 270 × 100 mm3 volume in 2.0 min, with 0.8 mm geometry resolution. We imaged eight body sites (Figure 1a) at seven timepoints spanning 172 days. The first imaging session (baseline, t0) was before the patient had developed any signs of cutaneous cGVHD. The remaining imaging sessions (t1–t6) captured cGVHD erythema. At each body site, a pair of photos was taken: cross-polarized (XP) that highlights subsurface features (e.g. hemoglobin that causes redness), and non-polarized (NP) that shows surface changes and texture (e.g. fine epidermal scale). One annotator manually cropped each body site of interest and manually registered the same body sites images of all timepoints. The total count was 112 3D-photos from the seven timepoints × eight body sites × two 3D-photos (cross- and non-polarized) per body site.
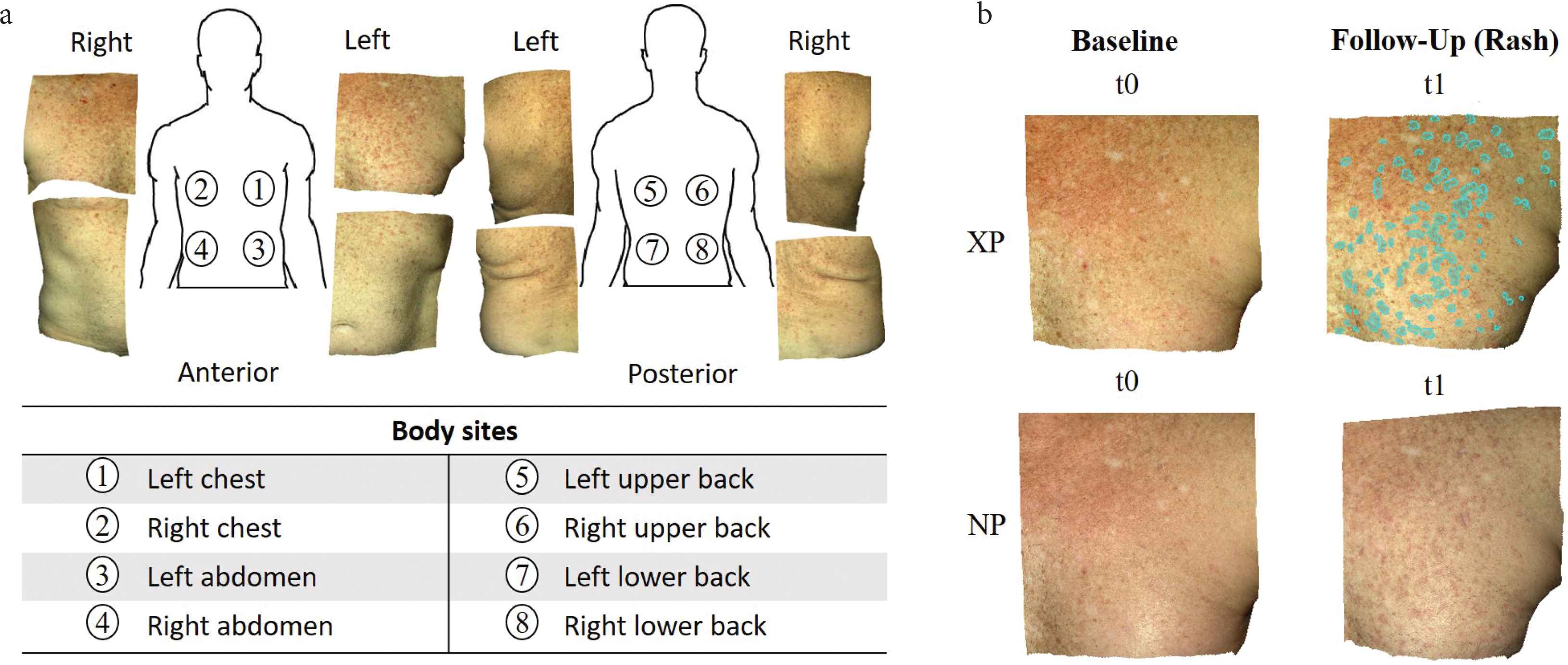
(a) Example of cross-polarized images of eight body sites from the first study session after baseline, which captured a new onset cutaneous chronic graft-versus-host disease rash. (b) The annotator simultaneously viewed four spatially registered skin photos: cross-polarized (XP) and non-polarized (NP) photos at baseline (no rash, t0) and at a follow-up session with a rash (in this example, t1). Annotations (cyan overlay) were done in the XP photo of the follow-up session (t1). Example shows “Do Not Miss” (DNM) annotations on left chest.
2.2. Human Annotation Methods: Traditional and Novel Confident
To develop an accurate computer-aided skin change algorithm, we proposed a new annotation approach: “Do Not Miss”/“Do Not Include” (DNM/DNI) segmentation. Two annotators who were trained by a board-certificated dermatologist independently marked each image by each annotation method: traditional and DNM/DNI segmentation. In traditional segmentation, annotators marked all areas that they felt are more likely to be active cGVHD erythema than inactive disease or normal skin. This traditional approach results in annotations of limited confidence that exactly divide the image into marked regions that are felt to be active erythema, and unmarked regions that are felt not to be active erythema. In contrast to this single marking strategy, DNM/DNI segmentation has annotators marking two types of areas, in which they have high confidence in the assignment: (i) definitely-affected areas as DNM and (ii) definitely-unaffected areas as DNI. Anything not marked in one of these two areas is not taken into account and, so, the annotator is not required to commit to presence or absence of active disease. In marked DNM/DNI areas, the annotator was asked to have sufficiently high-level confidence that an automated algorithm should always be penalized for missing any DNM pixels, and should always be penalized for including DNI pixels. During annotation, each annotator simultaneously viewed four images: the XP/NP image pair at baseline (t0) and the XP/NP image pair at a follow-up session (t1–t6, Figure 1b). Annotations were done on the XP image of each of the six follow-up sessions (t1–t6) and each of the eight body sites. This resulted in 48 XP images with traditional segmentation and 48 XP images with paired DNM and DNI segmentations. The two annotators were blinded to each other’s results.
2.3. Computer-Aided Algorithm to Detect Skin Change
We used an existing, pre-trained neural network [10,11] to extract features at each pixel of each image. Without adjusting any weights in the network, we designed an algorithm to detect skin changes based on the differences in features from the baseline photo to the time of rash. At each image pixel, the algorithm outputs a number on the scale of 0–1 that represents the difference in skin features between baseline and cGVHD rash photos (Figure 2). One image pair (left chest timepoints t0–t1) was used to fix the three algorithm parameters, and the remaining 47 image pairs were used to test the algorithm performance. Full algorithm details are provided in Supplementary Material 1.
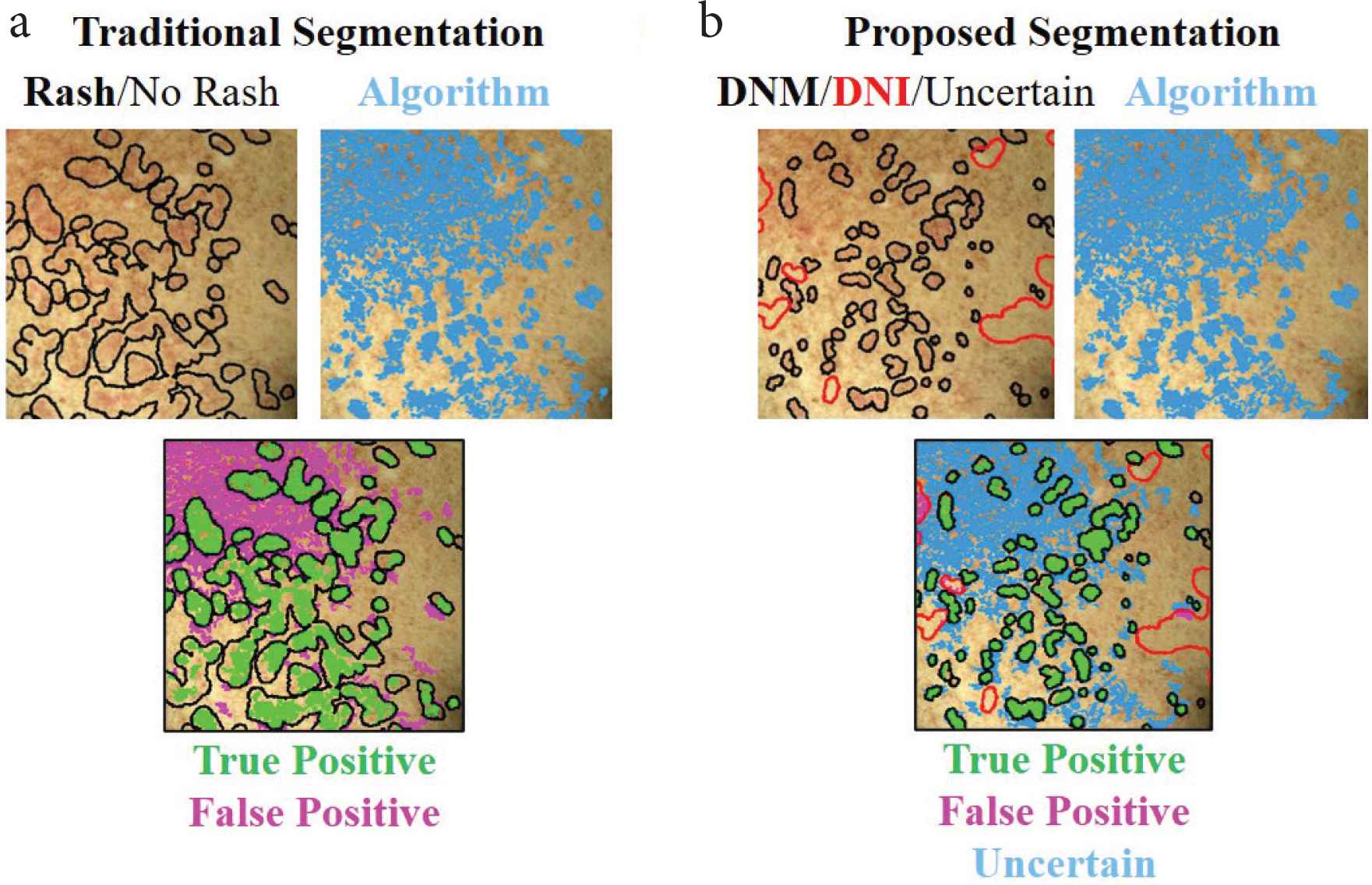
Algorithm outputs of detected skin change (blue areas) based on (a) traditional (black outline) and (b) proposed DNM/DNI (black/red outlines) segmentation methods. Whereas in traditional segmentation unmarked pixels represent “no rash”, in DNM/DNI segmentation the unmarked pixels are deemed uncertain by the annotator. Therefore, in the DNM/DNI method, the algorithm is not penalized for incorrectly detecting pixels that the human annotator was uncertain about. This results in less false positives (magenta), less true positives (green) areas, and uncertain pixels, compared to traditional segmentation method. DNM: “Do Not Miss”; DNI: “Do Not Include”. Example shows left chest of the first follow-up session.
2.4. Evaluation of Algorithm Performance in Active Erythema by Two Dermatologists
To test algorithm performance, two board-certified dermatologists independently evaluated each algorithm output. Each dermatologist independently scored the output in representing active erythema as 0 (poor: algorithm segmentation requires major revisions), 1 (good: requires minor revisions), or 2 (perfect: requires no revisions). They were blinded to the algorithm accuracy and annotations of the two annotators.
Each dermatologist did two types of evaluations: (#1) without viewing or (#2) while viewing the baseline skin photo. For evaluation #1, they simultaneously viewed the non-polarized and cross-polarized photos at a time of cGVHD rash. For evaluation #1, the four images were viewed (Figure 3a): a non-polarized (NP), and cross-polarized (XP) raw image, and the same images with algorithm output (blue overlay). For evaluation #2, the NP images were substituted for the baseline (pre-rash) XP image (Figure 3b). Typically, dermatologists would rapidly toggle their screen to display the raw photos and the algorithm-marked photo to enable a rapid short-term visual memory decision.
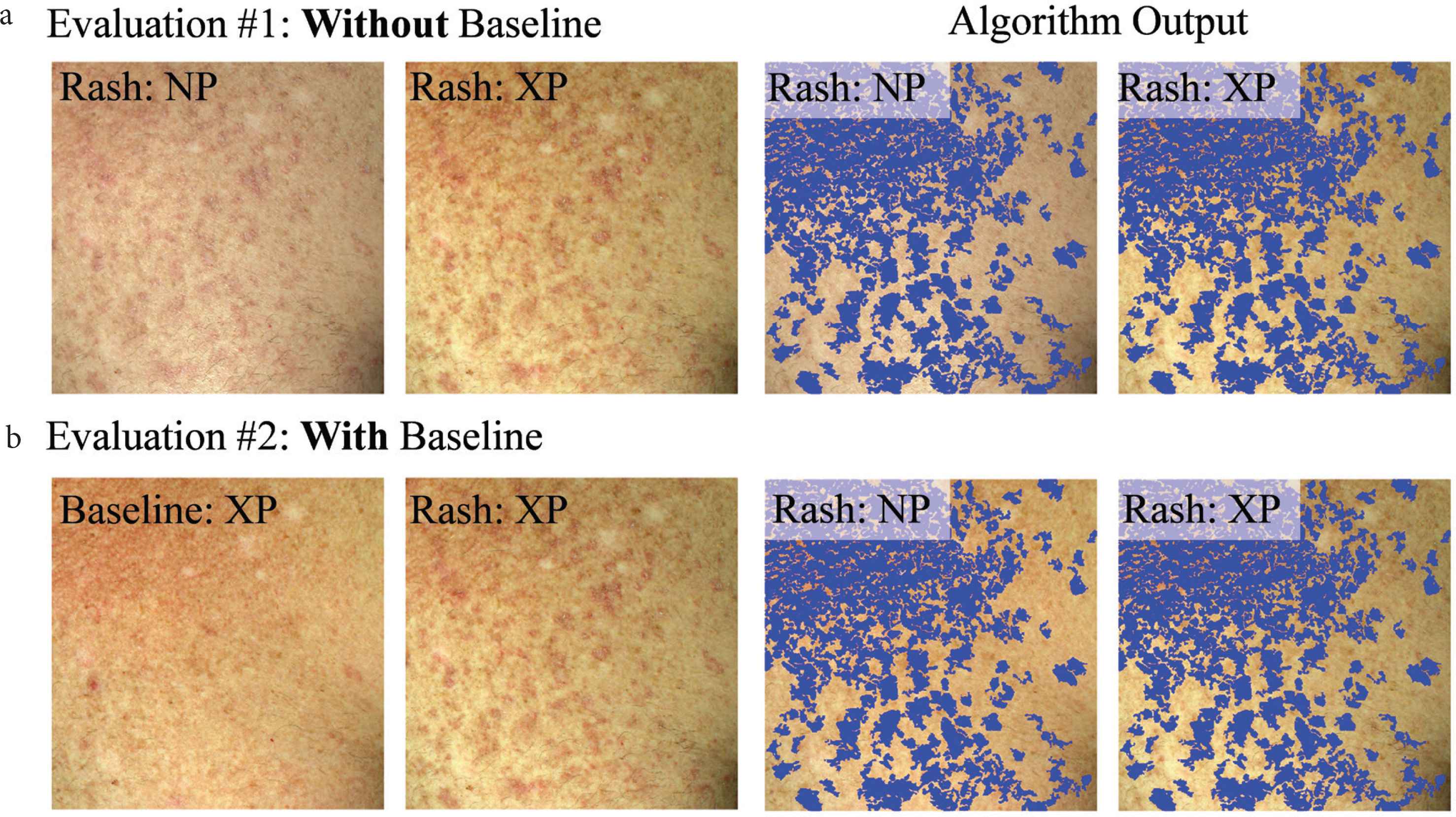
Example dataset that each dermatologist viewed to evaluate how well the algorithm output reflects active erythema (a) without viewing (evaluation #1) or (b) while viewing (evaluation #2) the baseline skin appearance photo. Algorithm output (blue overlay) are areas of skin change, detected by the developed computer-aided algorithm.
For each of the two evaluations, scoring was repeated in two rounds. The second round was done after evaluating the results of the first round and a 1 week washout period. In each round, each dermatologist evaluated all 47 image pairs in a random order unique to each dermatologist. Images were in a new random order in the second round. This resulted in each dermatologist scoring all 47 images four times.
2.5. Statistical Analysis
We first evaluated the accuracy of the change detection algorithm by using traditional human segmentations as the ground truth. Then, we independently evaluated the accuracy again based on the novel DNM/DNI segmentations. For each pixel, the algorithm output (skin change/no skin change) is compared to the ground truth in the corresponding human annotation (rash/no rash). When comparing algorithm output to traditional annotation, marked pixels represent “rash” and unmarked pixels represent “no rash”. In DNM/DNI annotation, DNM pixels represent “rash”, DNI pixels represent “no rash”, and unmarked pixels are uncertain and therefore are not compared to algorithm output. Accuracy = (TP + TN)/(TP + FN + TN + FP). True positive (TP) pixel represents algorithm and human agreeing on a skin change/rash. True negative (TN) pixel represents algorithm and human agreeing on no skin change/no rash. False positive (FP) pixel represents the algorithm detecting a skin change and the annotator assigning it as “no rash”. False negative (FN) pixel represents the algorithm not detecting skin change and the human assigning it as a “rash”.
To further evaluate algorithm performance, we calculated the average dermatologist score per two dermatologists, per each of the 47 XP/NP image pairs, resulting in average scores 0, 0.5, 1, 1.5, and 2. We used the Dice index to test the reliability between segmentation methods (DNM/DNI versus traditional) and algorithm outputs (DNM/DNI-based versus traditional-segmentation based skin change areas). The Dice index is the most widely used metric in medical imaging to evaluate the reliability of a segmentation method. It ranges from 0 (no overlap) to 1 (perfect overlap). The best algorithm for melanoma detection from dermoscopic images achieved an average Dice index of 0.85 in the 2017 International Skin Imaging Collaboration challenge [12].
To evaluate the intra-rater reliability of each dermatologist and the inter-rater reliability between the two dermatologists, we used intraclass correlation coefficients (ICCs). For ICC, we used the Eliasziw’s simultaneous random effects, absolute agreement, single-measure model [13]. Lower bound (single-sided) 95% confidence intervals were calculated using the corresponding “relInterIntra” function of the irr package in R [14]. Landis and Koch criteria were used for ICC interpretation, where values of 0.21–0.40 represent fair agreement, 0.41–0.60 moderate, 0.61–0.80 substantial, and 0.81–1.00 almost perfect agreement [15].
We used standard error of measurement (SEM) as a metric to evaluate variability among dermatologists’ scores of algorithm performance. SEM is interpreted as the assessment of reliability within individual subjects and has the same units as the measurement. The greater its value, the lower the reliability of the measurement [16]. Intra-rater SEMs summarize the variability inherent within the raters’ own measurements. Inter-rater SEMs include both the variability among raters’ measurements and the variability within raters’ measurements [17].
3. RESULTS
3.1. Improved Reliability for DNM/DNI Compared to Traditional Segmentation
We illustrate a segmentation approach (DNM/DNI) in which human annotators are confident in their markings of definitely affected and definitely unaffected areas of cutaneous cGVHD rash. DNM guides the algorithm to select pixels that should not be missed and DNI guides the algorithm to exclude pixels that should not be selected. Example annotations by two trained annotators are visualized in Figure 4.
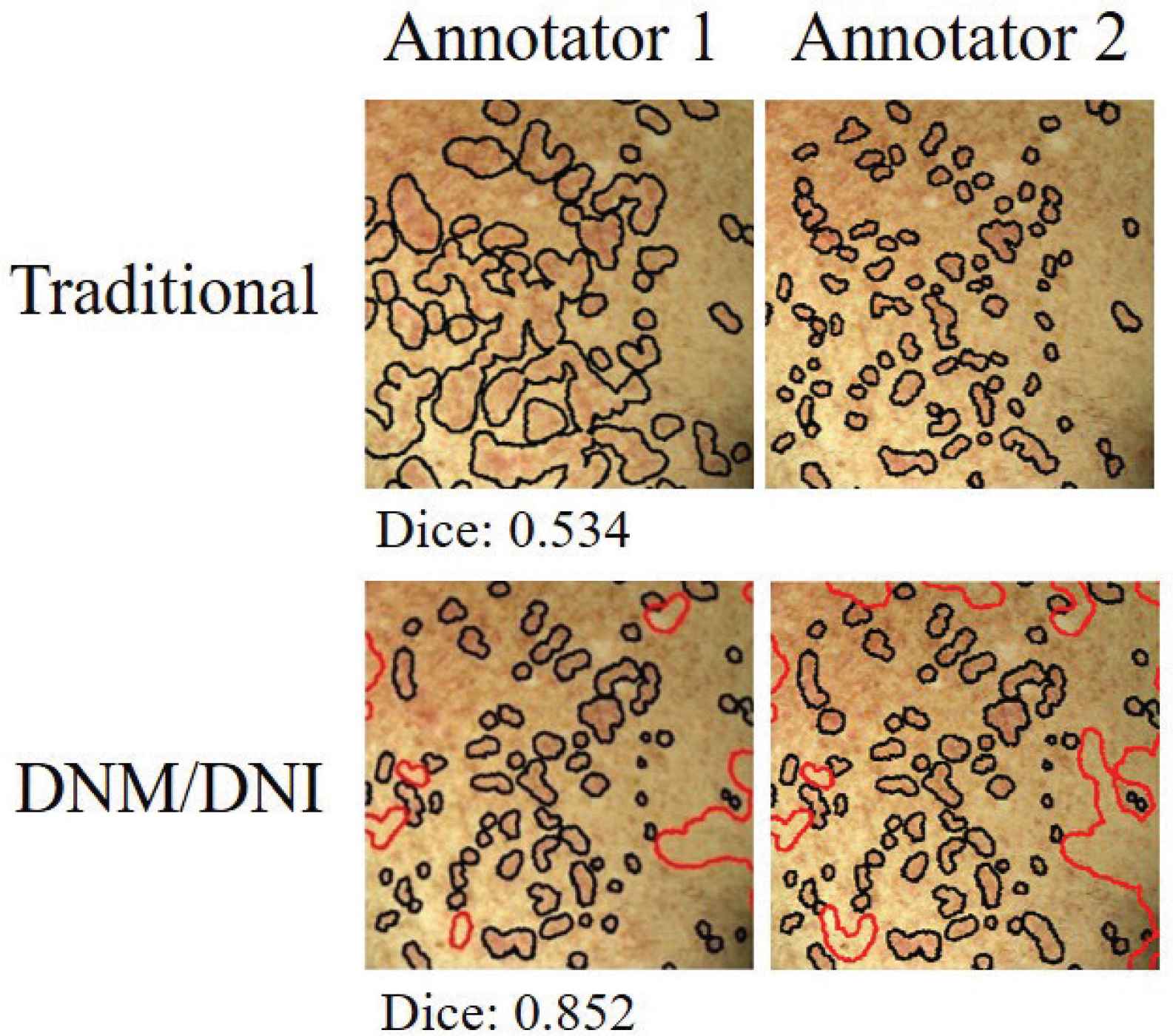
Independent markings by two annotators had higher agreement (Dice index of 0.852 versus 0.535) when using the “Do Not Miss/Do Not Include” (DNM/DNI) approach, compared to the traditional method. The Dice index measures the agreement between the two annotators on a scale of 0–1. For the DNM/DNI method, Dice is calculated by using both the DNM (black) and DNI (red) areas. Example shows left chest of the first follow-up session (t1).
The DNM/DNI segmentation approach achieved higher inter-rater reliability between annotators (median Dice index: 0.53–0.70) than traditional segmentation (0.36–0.47). The developed segmentation approach resulted in higher median Dice indices for algorithm output (0.94–0.96) than the traditional segmentation approach (0.73–0.81, Figure 5).
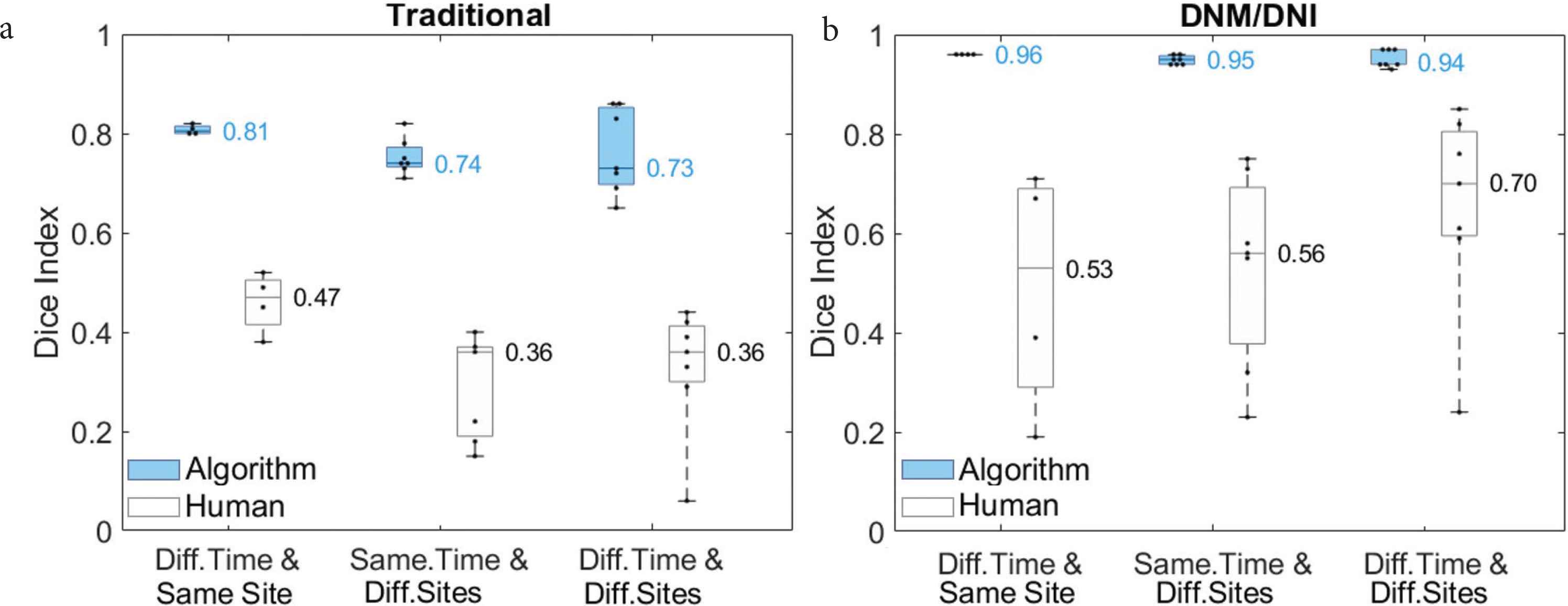
Compared to (a) traditional segmentation, (b) “Do Not Miss/Do Not Include” (DNM/DNI) segmentation resulted in higher average Dice indices between two human annotators (white) and between two algorithm outputs (blue). Each of the two algorithms was fixed on one of the annotators’ segmentations of the left chest at the first follow up t1. The algorithms were first tested on the left chest at t2–t5 (Different Timepoints and Same Site). They were then tested on the seven other body sites at t1 (Same Timepoint and Different Sites), and at follow-ups t2–t5 (Different Timepoint and Different Sites). Medians are shown next to the corresponding horizontal line in the boxplot.
3.2. Computer-Aided Skin Change Detection Algorithm: Tested on Cutaneous Chronic Graft-Versus-Host Disease
We demonstrate the ability to detect cutaneous cGVHD by evaluating change from a baseline skin photo. After fixing the algorithm with one of 48 images at the time of rash, the algorithm correctly assigned approximately 80% of the pixels in the 47 unseen images. Similar high performance was achieved both with traditional (median accuracy: 0.77, interquartile range 0.62–0.87) and the new DNM/DNI annotation method (0.81, 0.65–0.89, Figure 6).
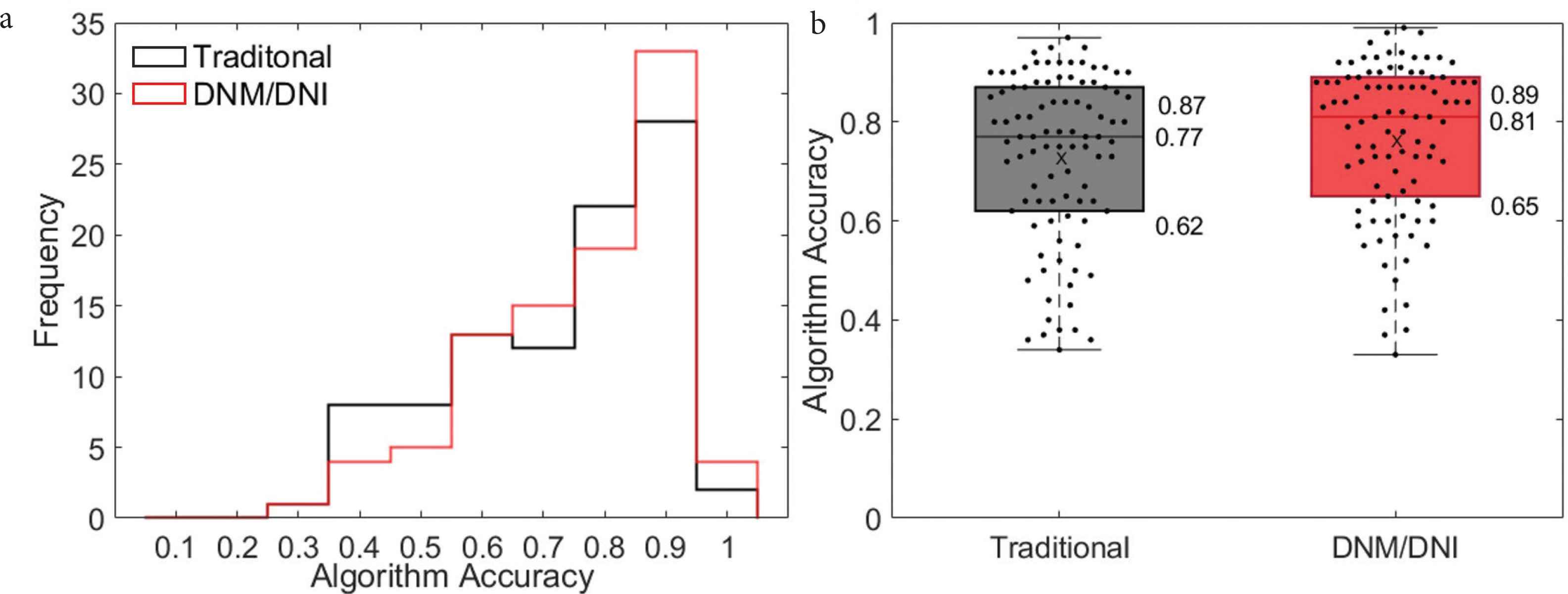
(a) Histogram and (b) boxplot show a similar algorithm accuracy across 47 test cGVHD images, regardless of whether the algorithm was based on traditional (black, median accuracy: 0.77, interquartile range: 0.62–0.87) or DNM/DNI annotations (red, 0.81, 0.65–0.89). Accuracy was calculated as the number of pixels that the algorithm correctly assigns, based on human segmentation. “X” is the average algorithm accuracy on the 47 images annotated by two annotators: 0.73 (traditional) and 0.77 (DNM/DNI).
3.3. Dermatologists’ Score on How Well Algorithm Detects Active Erythema
Two board-certified dermatologists evaluated how well the algorithm detects active erythema without viewing (evaluation #1) and while viewing (evaluation #2) baseline skin photos. In evaluation #1, they scored each output by simultaneously viewing the follow-up non-polarized and cross-polarized photos of a cGVHD rash and algorithm output (Figure 3a). In evaluation #2, they scored each output by simultaneously viewing cross-polarized photos of the baseline skin appearance and follow-up photo with a cGVHD rash, and algorithm output (Figure 3b). When viewing baseline photos, the dermatologists’ scoring of the algorithm was higher for those photos with higher algorithm accuracy. The average scores of the dermatologists correlated well with algorithm accuracy when viewing baseline photos, but not without viewing baseline photos (Figure 7).
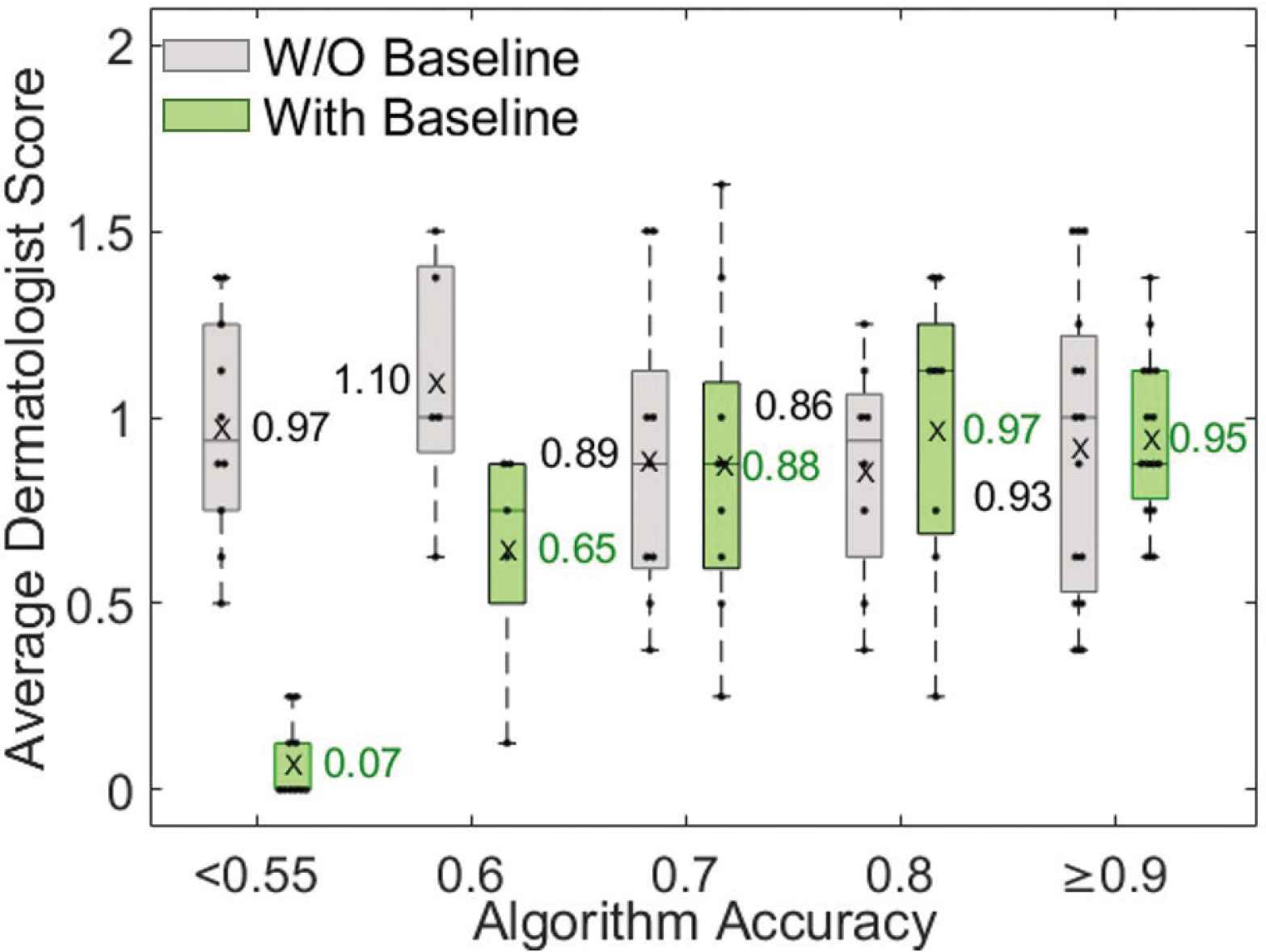
Dermatologist evaluation of how well the algorithm output identifies active erythema for photos with various algorithm accuracies. Dermatologists scores correlated with algorithm accuracy only when viewing baseline photos (green, evaluation #2). The average dermatologist score is the mean of four scores: two board-certified dermatologist scores in two rounds.
3.4. Viewing Baseline Photos Improves Reliability among Dermatologists
Viewing baseline skin photos (before the development of a cGVHD rash) improved the intra- and inter-rater reliability of the two dermatologists. The intra-rater ICC increased from a ‘fair’ value of 0.17 (95% confidence interval lower bound: −0.27) without viewing baseline photos to a ‘moderate’ value of 0.62 (0.33) while viewing baseline photos. The inter-rater reliability similarly increased from a ‘fair’ ICC of 0.19 (0.06) to a ‘moderate’ 0.51 (0.35) (Table 1) [15]. The intra-rater SEM improved from 0.52 to 0.36 and the inter-rater SEM improved from 0.51 to 0.40 with viewing baseline photos (Table 1).
| W/O baseline | With baseline | ||||
|---|---|---|---|---|---|
| Lower bound (95%) | ICC | Lower bound (95%) | ICC | ||
| ICC | Intra | −0.27 | 0.17 | 0.33 | 0.62 |
| Inter | 0.06 | 0.19 | 0.35 | 0.51 | |
| SEM | Intra | 0.52 | 0.36 | ||
| Inter | 0.51 | 0.40 | |||
Viewing baseline photos increased intra- and inter-rater reliability (intraclass correlation coefficient, ICC). Both the 95% lower bound of one-sided confidence interval and point estimate are shown. Baseline photo viewing also decreased intra- and inter-rater variability (standard error of measurement: SEM)
4. DISCUSSION
Longitudinal tracking of erythema is critical for treatment decisions in patients with cutaneous cGVHD and would benefit greatly from automated image analysis methods. We designed an algorithm to detect skin change based on a pre-trained neural network as a feature extractor. Our algorithm did not adjust or train the network but rather used a human annotation of a single rash photo to fix three algorithm parameters to detect changes from the patient’s baseline before rash. Our pilot study confirmed our hypothesis that asking humans to only mark parts of image in which they are confident improves both human and algorithm reliability. Further, we found that viewing photos of baseline skin appearance improves dermatologists’ reliability.
To calculate the severity of GVHD, the European Society for Blood and Marrow Transplantation has developed an eGVHD app. However, eGVHD does not address longitudinal patient evaluations [18]. In dermatology, longitudinal tracking of suspicious pigmented lesions via short-term digital dermoscopy has dramatically increased the accuracy of melanoma detection [19]. In radiology, an automated change detection system was shown to outperform experts when assessing multiple sclerosis progression in magnetic resonance scans. Whereas experts missed 58% of cases with progressing disease, the algorithm missed 5% [20]. Similarly, longitudinal computer tomography of the liver has been shown to enhance detection of tumors [21]. To the best of our knowledge, no algorithms exist for detecting change in cutaneous cGVHD or other inflammatory skin diseases.
Uncertainty is a common challenge in medical image segmentation due to poor contrast or other restrictions imposed by the image acquisition or variations in annotation between experts. In magnetic resonance and computed tomography images, accuracy of automated segmentation algorithms has been improved by accounting for the uncertainty in learned model parameters [22,23]. Instead of embedding uncertainty in the algorithm development, we explored a more confident annotation approach. We developed a DNM/DNI segmentation method where the annotator is asked to only mark areas in which they are confident. This is a major departure from the traditional approach where the annotator is asked to assign all parts of the image as affected or non-affected. Although our change detection algorithm achieved a similar accuracy (>0.7), regardless of which human annotation method it was based on, the DNM/DNI approach resulted in a higher inter-rater agreement (Dice: 0.70 versus 0.47) than the traditional method. DNM/DNI may aid in the development of more accurate and reliable segmentation algorithms in a variety of medical imaging applications beyond inflammatory skin disease.
Although baseline photography is commonly used to detect change in pigmented lesions [8,9], its value is unknown in detecting change in an inflammatory disease. Individual processing of a rash photo without baseline skin appearance may not account for other causes of redness, such as sun-damaged skin or an underlying dermatologic condition. Furthermore, detection of erythema in patients with darker skin remains a challenge both by visual inspection, as well as by computer-aided methods. Without viewing baseline skin photos, in-person assessments of the body surface area affected by cGVHD erythema have achieved modest (ICC: 0.41–0.60) inter-rater agreement [5] by Landis and Koch criteria [15]. In a previous study [7], we tracked erythema at each imaging session over time, without taking into account skin appearance at baseline. Six trained raters, while only viewing the rash photo, achieved poor agreement in the assessment of body surface area in those 3D photos (ICC = 0.09). Consistent with these prior studies, we found poor agreement (0–0.20) in the evaluation of algorithm output when the dermatologists did not view a photo of baseline skin appearance. By contrast, dermatologist intra- (0.62) and inter-rater (0.51) agreement improved to modest when they viewed baseline photos. Viewing baseline skin photos improved the agreement between dermatologists on algorithm performance (0.51 versus 0.19). Capturing baseline photos in research studies and clinical practice may increase the accuracy in assessing skin change, decrease the disagreement between raters and improve the consistency among expert clinicians to identify areas of active disease.
Our study has several limitations. The algorithm performance relies on a precise manual alignment of longitudinal skin photos of the same body site. Because human annotators only marked areas that they were confident about as either affected or unaffected, the developed algorithm output of skin change included large areas of uncertainty. Rather than detecting cGVHD rash based on information in a single image pixel, future studies could explore automated detection of cGVHD rash based on the information available in the image as a whole (all pixels). We tested the developed algorithm on a single patient’s data with a papulosquamous rash of cGVHD, therefore the algorithm did not have to account for the pleomorphic presentations of cGVHD. We hope to address this in the future by expanding our dataset. If validated in a larger patient population, this method may assist clinicians in monitoring and managing cutaneous cGVHD.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
AUTHORS’ CONTRIBUTION
BMD and ERT secured funding, designed the study. KP collected patient photos. ERT provided ground truth markings on photos and trained all evaluators. XL, KP, IS, and TR performed data analysis. ADC performed statistical analysis. LEW and ERT scored the algorithm performance. IS, XL, and ERT wrote the manuscript. All authors have approved the final version of the manuscript.
ACKNOWLEDGMENTS
This work is supported by Career Development Award Number IK2 CX001785 from the United Sates Department of Veterans Affairs Clinical Science R&D (CSRD) Service, the National Institutes of Health Grant R21-AR074589, a Discovery Grant from Vanderbilt University, and Dermatology Foundation.
SUPPLEMENTARY MATERIAL
Supplementary data related to this article can be found at
Footnotes
REFERENCES
Cite this article
TY - JOUR AU - Xiaoqi Liu AU - Kelsey Parks AU - Inga Saknite AU - Tahsin Reasat AU - Austin D. Cronin AU - Lee E. Wheless AU - Benoit M. Dawant AU - Eric R. Tkaczyk PY - 2021 DA - 2021/07/15 TI - Baseline Photos and Confident Annotation Improve Automated Detection of Cutaneous Graft-Versus-Host Disease JO - Clinical Hematology International SP - 108 EP - 115 VL - 3 IS - 3 SN - 2590-0048 UR - https://doi.org/10.2991/chi.k.210704.001 DO - 10.2991/chi.k.210704.001 ID - Liu2021 ER -
