RadCloud—An Artificial Intelligence-Based Research Platform Integrating Machine Learning-Based Radiomics, Deep Learning, and Data Management
- DOI
- 10.2991/jaims.d.210617.001How to use a DOI?
- Keywords
- Radiomics; Machine learning; Neural network; Data management
- Abstract
Radiomics and artificial intelligence (AI) are two rapidly advancing techniques in precision medicine for the purpose of disease diagnosis, prognosis, surveillance, and personalized therapy. This paper introduces RadCloud, an artificial intelligent (AI) research platform that supports clinical studies. It integrates machine learning (ML)-based radiomics, deep learning (DL), and data management to simplify AI-based research, supporting rapid introduction of AI algorithms across various medical imaging specialties to meet the ever-increasing demands of future clinical research. This platform has been successfully applied for tumor detection, biomarker identification, prognosis, and treatment effect assessment across various image modalities (MR, PET/CT, CTA, US, MG, etc.) and a variety of organs (breast, lung, kidney, liver, rectum, thyroid, bone, etc). The proposed platform has shown great potential in supporting clinical studies for precision medicine.
- Copyright
- © 2021 The Authors. Published by Atlantis Press B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Radiomics is a quantitative image analysis technique through extraction of quantitative features from medical images [such as magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), ultrasound (US), etc.] [1], aiming to relate large-scale data mining of images to clinical and biological endpoints [2]. Radiomics has been widely applied to many different precision medicine applications across a variety of organs and imaging modalities, with an intent to provide assistance for disease screening, diagnosis, surgery, and outcome prediction, etc. [2].
Recently, artificial intelligence (AI) has been coupled with radiomics for oncological and nononcological applications [3,4], due to its greater capability in handling massive amount of data compared to the traditional statistical methods. AI-based radiomics has been increasingly exploited for clinical studies with ever-developing advances in computational power, availability of large datasets, and machine learning (ML)/deep learning (DL) algorithms. It can assist radiologists and clinical researchers in reducing the heavy workload and providing opportunities to deal with more complex and sophisticated radiological problems [5].
However, implementing AI-based radiomics into clinical practice requires conquering some technical problems [3] such as the time-consuming workflow, uncertain reproducibility of results among different scanners, acquisition protocols, and image-processing approaches, regulatory issues concerning privacy and ethics, and data protection, etc. Moreover, radiologists and clinical researchers are often not experts in data science and/or AI algorithms, and a knowledge gap exists between information extraction and understanding of meaningful clinical data, which will limit clinical researchers for taking full advantages of AI benefits in radiomics studies. Based on status quo thereof, RadCloud (http://119.8.187.142/), an AI research platform is developed to support studies with integrated data management, ML-based radiomics, and DL algorithms.
The remaining parts of this paper are organized as follows: Section 2 describes the proposed research platform for radiomics studies; Section 3 reviews latest applications of the proposed platform; and Section 4 concludes the contributions and provides the future work for improvement.
2. RADCLOUD RESEARCH PLATFORM
RadCloud is a research platform supporting both ML-based radiomics analysis and DL-based medical image analysis, specialized in managing, analyzing, and mining image data from medical examination and clinical reports. The platform aims to implement a whole data science research life cycle to provide an All-in-One data-driven solution to radiomics studies for clinical researchers with a simple user-interface (UI) from data collection, annotation, feature extraction to model training, validation, and testing. The RadCloud platform offers a project management approach where clinical researchers can set multi-level projects, ranging from individual projects, department projects, institute projects to multi-center projects. The isolation of project access protects creative ideas whilst keeping track of the research progress. The high availability enabled by the browser/server (B/S) architecture makes the platform easy to use and access. In the meantime, with high concurrency powered by flexible storage and balanced load, the platform can meet the needs for large projects.
2.1. AI Research Unit
The AI research unit integrates data annotation, ML-based radiomics, and DL-based medical image analysis module.
2.1.1. Data annotation module
The data annotation module provides numerous manual and semi-automatic annotation tools to support various annotation tasks such as segmentation, classification, target detection, key point positioning, etc., along with customizable multi-level tags for annotation content management and a well-equipped multi-blind review mechanism. The annotation toolbox provides common annotation functions such as stipple, line, ellipse, polygon, rectangle, key point, classification label, and measurements, and more advanced functions such as brush, magic wand, curve sketching, contour instance measurements, etc. It also comes along with semi-automatic annotation tools, including adaptive fitting, interpolation and Lasso tool. Thus, users do not need to switch to other annotation software. Figure 1 shows the UI of data annotation tools for region of interest (ROI) segmentation.
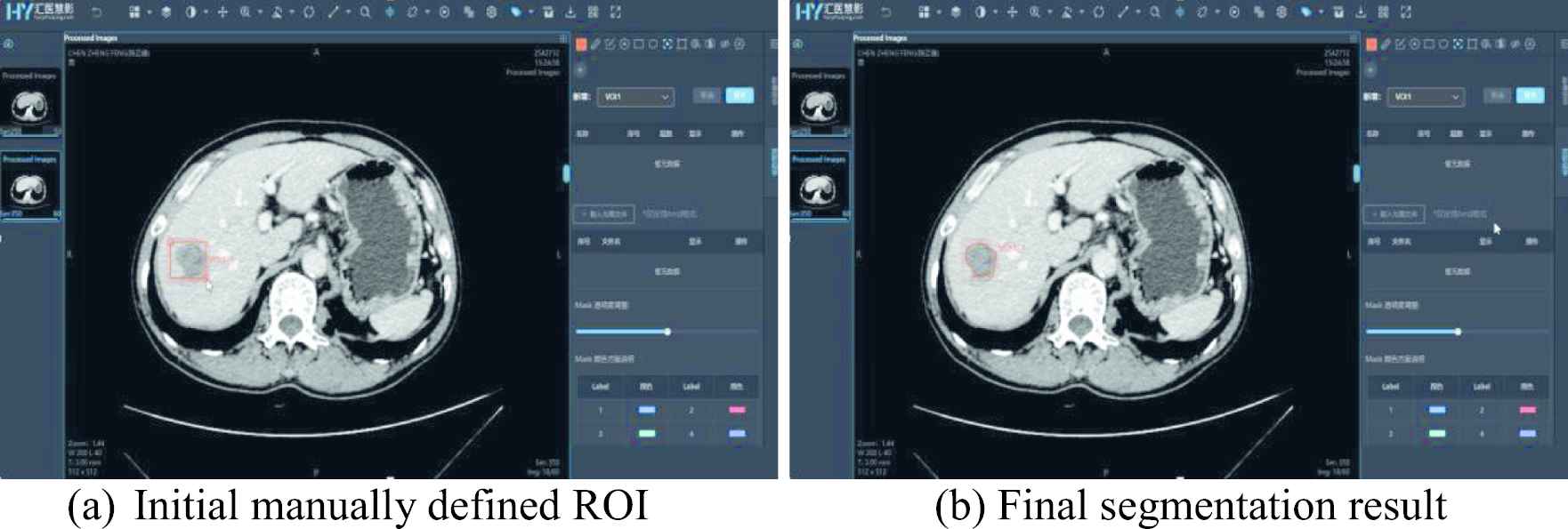
Semi-automatic ROI annotation.
This module also supports data management. It supports a batch upload of data with breakpoint continuation and a direct connection to imaging equipment/PACS, followed by the cloud distributed storage with a build-in desensitization tool for the anonymization of personal data, such as patient's name, institute's name, etc. After being uploaded, the original data including PET/CT/MR/DR/US images and structured clinical information (diagnosis results/pathological grades/gene protein) data can be grouped by area and disease type, and the records of the uploading, distribution, and the utilization history are kept for future references. The data is managed according to the projects, advanced users can allocate data to multi-level projects and set review permissions within the platform, with isolated access and real-time record of the annotation progress. For an individual project, annotation can be performed from different devices and stored securely, while a database is shared to make research/communication in a common annotation environment for a collaborative project.
After an annotation task is completed, annotated data can be assigned to reviewers for assessment. The reviewers can approve or modify the annotation results. Final annotation and reviewers' marking results are mutually inferable and traceable, which forms a flexible mechanism for double-blind and multi-blind annotations. The corrected annotated data after reviewing can be set as gold-standard annotations by administrator to be used for further research or education purposes.
The annotation module can prevent the loss/damage of annotated data by hosting the service on cloud, allowing efficient co-operative annotation without the limitations of computer systems and the restriction of tools. Its data management function enables the management of project groups, data, and collaborations, and ensures the communication work in a safe and secure cyberspace. The customization option for annotation tags suiting for various clinical scenarios can satisfy the demand for flexibility.
2.1.2. ML-based radiomics module
After the data are annotated, the radiomics module will carry out feature extraction and feature selection and ML-based prediction. Figure 2 demonstrated the pipeline of this module.
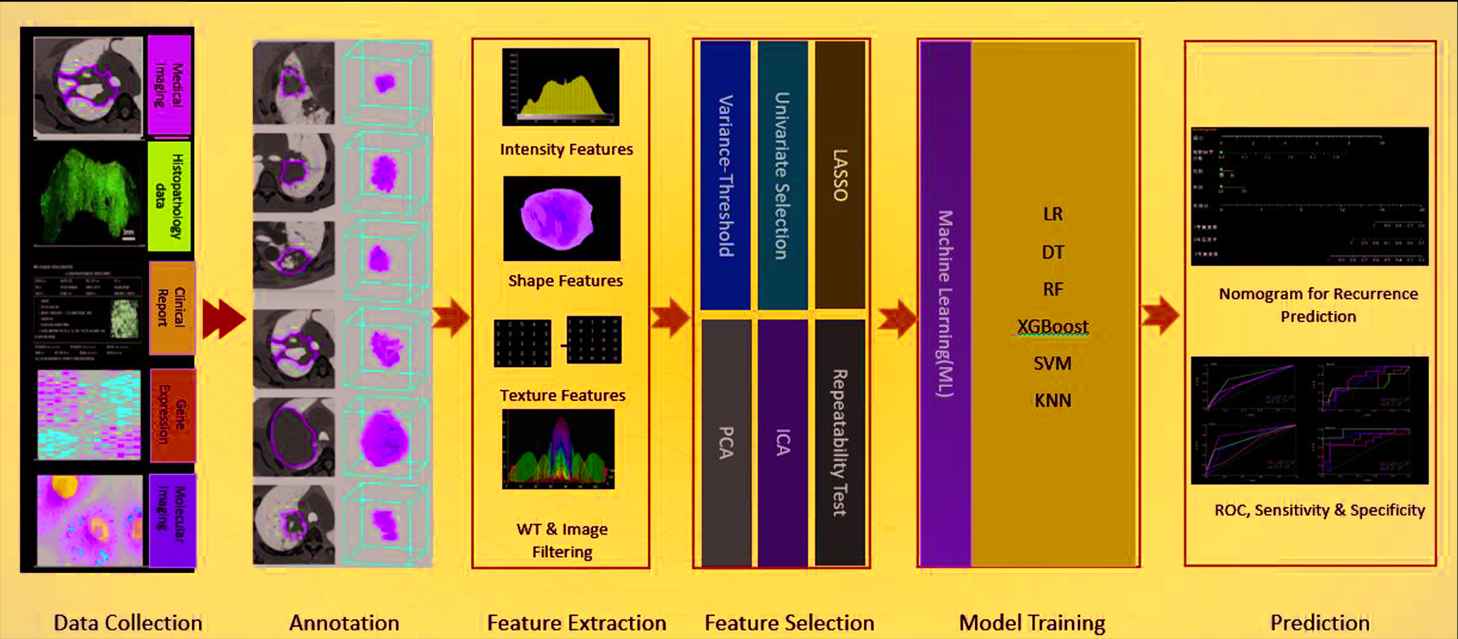
An overview of the ML-based radiomics workflow.
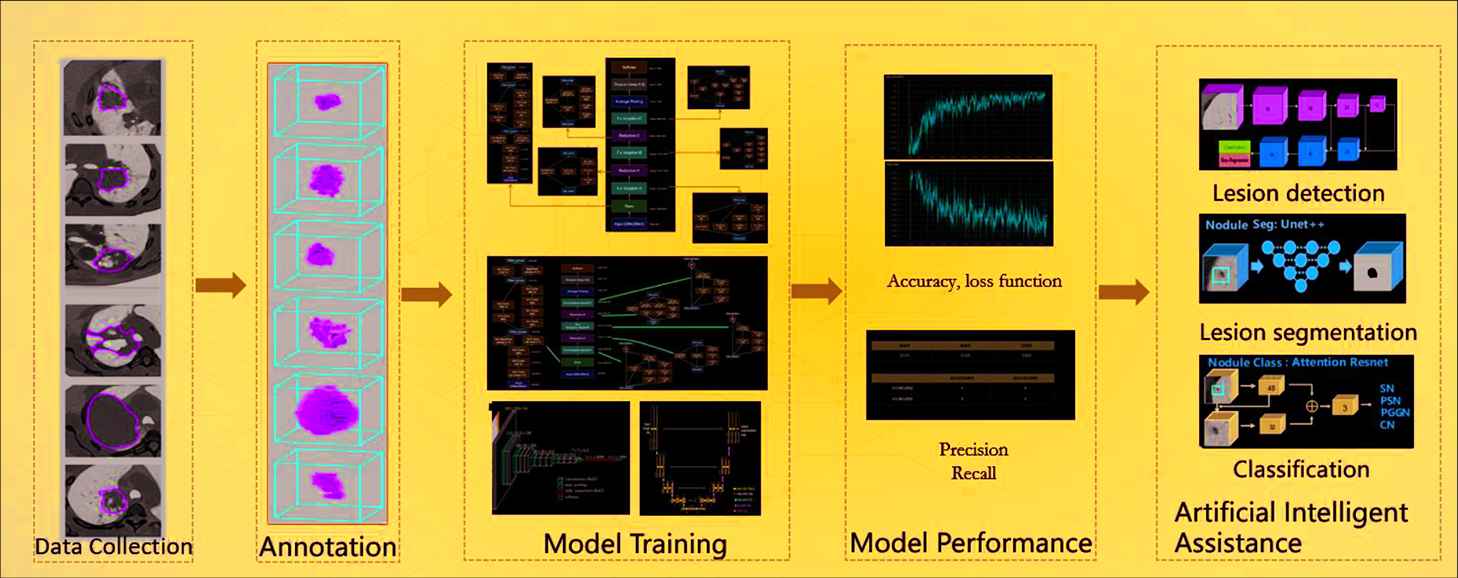
A workflow of deep learning medical image analysis.
Feature Extraction
In the proposed platform, 1,400+ features can be automatically extracted from annotated images, such as
First-order statistics including mean, standard deviation, variance, maximum, median, range, kurtosis, etc.
Texture features including gray level co-occurrence matrix (GLCM), gray level size zone matrix (GLSZM), gray level run length matrix (GLRLM), etc.
Shape features including volume, surface area, compactness, maximum diameter, flatness, etc.
High-order features including first-order statistics and texture features after exponential, square, square-root, filtering, etc.
Most features shown above are in compliance with feature definition as described by the Imaging Biomarker Standardization Initiative (IBSI), which is an independent international collaboration for standardizing the extraction of image biomarkers (features) from acquired imaging and improving reproducibility for the purpose of radiomics [6]. Users can also define customized features.
Feature Selection
The feature selection module is used for feature selection or dimension reduction to improve the accuracy or boost the performance of modellng. It includes variance threshold for low variance feature removal, SelectKBest, Lasso (least absolute shrinkage and selection operator) for sparse feature selection. These methods are all based upon Python machine leaning package sklearn (www.scikit-learn.org). Independent component analysis (ICA) for separating multivariate signal, principal component analysis (PCA) for dimension reduction, correlation analysis, clustering analysis can also be performed to validate the feature selection.
ML Model Setup
The ML module has implemented a variety of ML models, including linear regression (LR), decision tree (DT), radom forest (RF), extreme gradient boosting (XGBoost), support vector machine (SVM), K-nearest neighbors (KNN), and allowed relative hyperparameters adjustment. Nomogram model is also included in this module to facilitate the combination analysis of radiomics and clinical factors.
2.1.3. DL-based medical image analysis module
The DL-based medical image analysis module provides a much easier way for medical image analysis. Different from ML-based radiomics, which extracted predefined features, DL approaches can extract more complicated and high-dimensional features from multi-layer of neural networks. Figure 3 illustrates a workflow of DL-based analysis. The DL models include segmentation models (DeepLab, U-Net, Mask-RCNN), detection models (YOLO, FPN, ResNet series and Faster-RCNN), and classification models (Inception, VGG). The DL module mainly involves the following eight steps:
Dataset collection: upload annotated/un-annotated medical images and related clinical information to the platform;
Data annotation: annotation module described in Section 2.1.1;
Data split: training, validation, and testing dataset division at random or predefined groups;
Model selection: select a DL model from the model library of the platform for training from scratch, or importing internally pretrained and saved models or format-compatible external models;
Hyperparameter configuration: GPU index, epoch number, batch size, training times per epoch, optimizer, learning rate, loss function, anchor ratios if applicable, etc.;
Model training: perform and monitor model training through visualizing the loss function curvature and the landscapes of other indices during the training process;
Model test and validation: test and validate the model with predefined validation and test datasets;
Output: output the trained model in a given format (such as HDF5, JASON, YAML).
With 2 * GPU (1080 TI 12GB), 1 * CPU(intel Xeon(R)), training 100 samples(epoch = 50, batch = 4) takes 2–6 hours for classification task and 4–12 hours for segmentation and detection tasks also differs depending on the different frameworks applied.
3. APPLICATIONS OF THE RADCLOUD PLATFORM
The RadCloud platform has been used for tumor detection, cancer stage prediction, biomarker identification, prognosis, and treatment effect prediction across various image modalities (MR, PET/CT, CTA, US, MG etc) and a variety of organs (breast, lung, kidney, liver, rectum, thyroid, bone etc.).
3.1. ML-Based Radiomics Analysis
3.1.1. Lung
The feature extraction module in the RadCloud platform have been used for extracting more than one thousand radiomic features from 2D/3D ROIs of CT and PET images to improve the prediction of the lymphovasular invasive status and prognosis of patients with lung adenocarcinoma [7,8]. It was also used to extract radiomic features of lung tumors from CT component of PET/CT images for distinguishing benign and malignant lung lesions [9].
Radiomics-based predictive models with RF and KNN classifiers have been used for identifying glucocorticoid-sensitive patients with connective tissue disease (CTD)-related interstitial lung disease (ILD) for optimizing glucocorticoid treatment [10].
3.1.2. Breast
Following the workflow of radiomics analysis built in RadCloud, a multivariate logistic regression based radiomics model was developed to predict benign and malignant breast tumors (<1 cm) from contrast-enhanced spectral mammography (CESM) images [11]. The model showed a good prediction performance with an area under the curve (AUC) of 0.940 in a retrospective study of 139 patients. To identify whether the existence of HER-2 oncogene was associated with the poor prognosis and responses to certain chemotherapies (e.g., trastuzumab), the radiomics models with SVM and LR classifiers using mammography (MG) radiomics features, were developed and validated on an MG dataset of 526 patients [12]. In addition, a LR-based MR radiomics model was used for predicting the Ki-67 proliferation index in patients with invasive ductal breast cancer from MR images [13].
Similarly, radiomics models with three ML classifiers (LR, XGBoost, and SVM) were constructed based on dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) of primary breast cancer patients and used to predict the existence of axillary SLN metastasis, an important biomarker for determining the cancer stage and adjuvant treatment plan decision after surgery. The SVM classifier achieved the highest classification performance with an accuracy of 0.85 and an AUC of 0.83 in a clinical study of 62 patients [14].
3.1.3. Liver
To preoperatively differentiate focal nodular hyperplasia (FNH), the second most common benign liver tumor, from HCC in the noncirrhotic liver for facilitating clinical decision-making of treatment, a radiomics-based regression model was implemented and validated, in which the feature extraction module of RadCloud was used for radiomic feature extraction from CT images [15]. To predict microvascular invasion (MVI) of HCC, six radiomics-based classifiers were constructed. Among them, SVM, XGBoost, and LR classifiers showed great diagnostic performance with AUCs > 0.93 [16].
In addition, four radiomics models with ML classifiers (MLP, SVM, LR, and KN) supported by the RadCloud platform have been successfully established for the preoperative diagnosis of dual-phenotype hepatocellular carcinoma (DPHCC) and the prediction of patient prognosis from MR images [17]. Moreover, the radiomics-based classification of HCC and hepatic hemangioma (HH) using MR images showed a good performance equivalent to an experienced radiologist with 10 years experience.
3.1.4. Kidney
Clear cell renal cell carcinoma (ccRCC) is an extremely aggressive urologic neoplasm, and ccRCC patients with synchronous distant metastasis (SDM) have poor prognosis. Early identification of SDM can ensure the reasonable and effective treatment. In a retrospective study of 201 patients, the RadCloud platform was used to construct an MR radiomics model with multivariable logistic regression for the individualized identification of SDM in patients with ccRCC. The model achieved a convincing predictive performance with an AUC of 0.854 [18].
In addition, the RadCloud platform help build CT radiomics models for distinguishing benign rental angiomyolipoma without visible fat from malignant ccRCC [19], discriminating low-grade and high-grade ccRCCs [20,21], differentiating sarcomatoidrenal cell carcinoma (SRCC) and ccRCC [22], and distinguishing benign fat-poor angiomyolipoma (fpAML) from malignant chromophobe renal cell carcinoma (chRCC) [23].
3.1.5. Rectum
The RadCloud platform has been applied for rectal cancer (RC) studies. For example, in a retrospective trial of 166 consecutive RC cases, an XGBoost-based MR radiomics model was constructed to differentiate local recurrence of RC from nonrecurrence lesions at the site of anastomosis, with an AUC of 0.864 [24]. In a retrospective study of 165 consecutive patients with locally advanced rectal cancer (LARC), an LR-based MR radiomics model was developed to predict the pathological complete response (pCR) after neoadjuvant chemoradiotherapy (nCRT) in order to identify candidates for optimal treatments [25]. In a study of 61 patients with renal tumors, CT radiomics models with five ML classifiers were implemented for differential diagnosis of malignant renal chromophobe cell carcinoma (chRCC) and benign renal oncocytoma (RO) from CT images [26]. MR radiomics models with SVM and LRT were developed to differentiate benign rectal adenoma and adenoma with canceration [27].
The RadCloud platform has also been applied in grading tumor studies. MR radiomics models with different ML classifiers (such as MLP, LR, SVM, DT, RF, and KNN, etc.) were developed for predicting tumor stages simplified as T(T1-T2) and N(N1-N2) stages [28], and tumor regression grades (TRGs) for evaluating the treatment response of LARC after neoadjuvant chemoradiotherapy (nCRT) of RC [29].
3.2. DL-Based Medical Image Analysis Module
The DL-Based Medical Image Analysis Module in the RadCloud platform has started being used for clinical researches. For example, transfer learning by tunning pretrained deep convolution neural network (CNN) models (VGG16, ResNet 18, GoogleNet, inception-V3 and AlexNet) has been used for the classification of benign and malignant thyroid nodules based on hybrid features of conventional US and US elasticity imaging [32]. 1156 thyroid nodule US images in 233 patients were included in this study, resulting in 539 benign images and 617 malignant images in total [32]. U-net based deep CNN models integrated in DL module have been used for automatic segmentation of Type B aortic dissection (TBAD) from computed tomography angiography (CTA) [33]. In the study, the model was trained on CTA images of 276 patients with TBAD, and had an accuracy of a mean Dice similarity coefficient (DSC) of 0.93 (p < 0.05), and a segmentation speed of 0.038 ± 0.006 s/image. Figure 4 illustrates the UI of automatic segmentation and parameter measurements of Type B aortic dissection (TBAD) which has already been developed as a stand-alone AI-assisted software application.
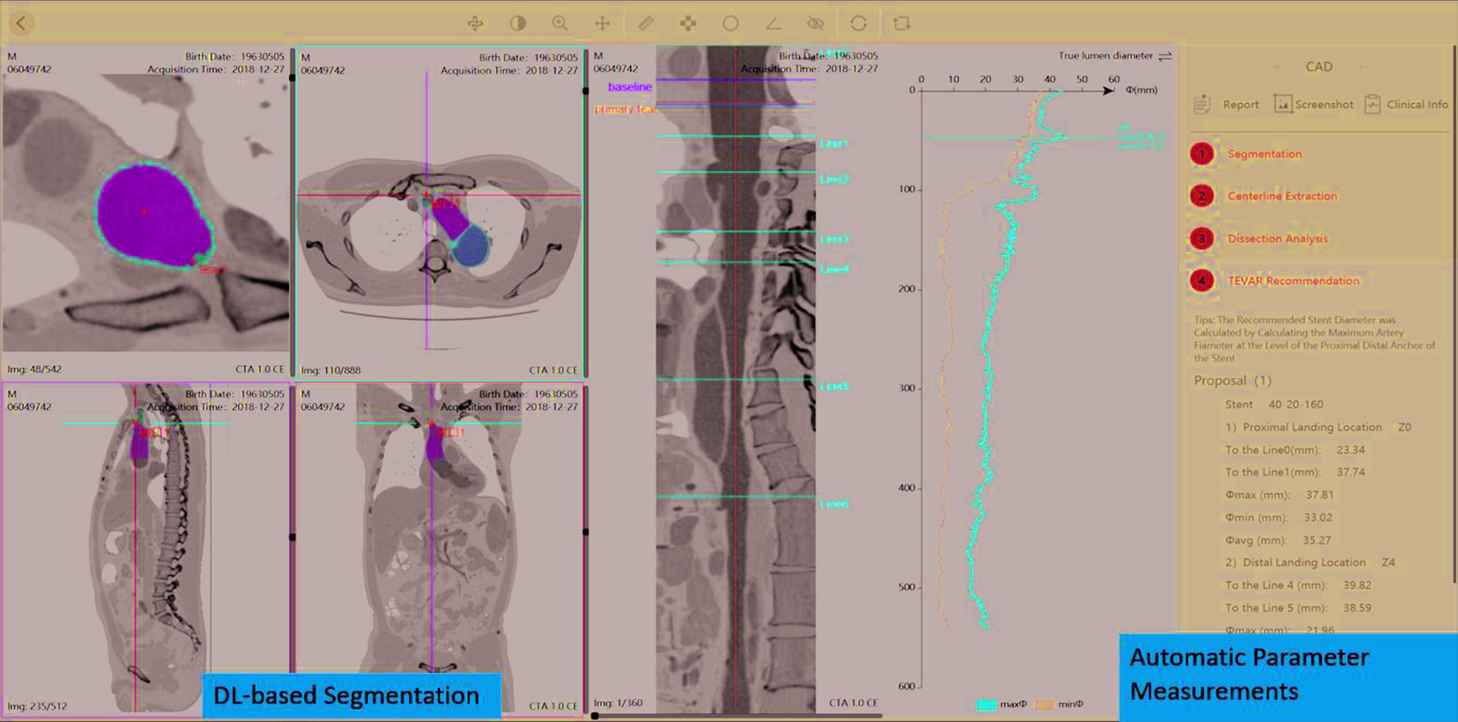
A user-interface of the RadCloud platform illustrating automatic segmentation and parameter measurements of Type B Aortic Dissection (TBAD).
Lung nodule detection with a novel fusion mechanism with results from four state-of-art object detectors including Retina, Retina U-net, Mask R-CNN, and Faster R-CNN+ was established under the help of this platform and had demonstrated best results in luna16 dataset [34]. More recently, U-net- based DL models trained on annotated datasets of COVID-19 have also been used for the potential lung lesion segmentation and affected volume measurement of both lungs, as well as COVID-19 probability prediction [35].
4. CONCLUSION
In this paper, we have presented an AI-based research platform and reviewed its applications. The platform integrates functions including ML-based radiomics analysis, DL-based medical image processing, and data management. The access control not only protects creative research ideas but also keeps the administrator aware of the team progress. Moreover, the strategy to host applications and service on cloud provides affordable, easy-to-use and remotely accessible data storage, and file sharing.
The ML-based radiomics analysis methods built in the platform have been successfully used for various clinical studies in oncology, the same goes for DL-based module. The proposed platform has shown great potential in supporting medical imaging studies for precision medicine. To sum up, RadCloud research platform provided a one-stop AI-assisted research tool for boosting scientific studies. from automatic radiomic feature extraction, ML setup and fine-tune, to button-initiated training, validation and test of selected CNN frameworks, hyper-parameter adjustment, aiming at a friendly user experience for nonprogramers.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
ACKNOWLEDGMENTS
Thanks to all participants in this study.
REFERENCES
Cite this article
TY - JOUR AU - Geng Yayuan AU - Zhang Fengyan AU - Zhang Ran AU - Chen Ying AU - Xia Yuwei AU - Wang Fang AU - Yang Xunhong AU - Zuo Panli AU - Chai Xiangfei PY - 2021 DA - 2021/06/21 TI - RadCloud—An Artificial Intelligence-Based Research Platform Integrating Machine Learning-Based Radiomics, Deep Learning, and Data Management JO - Journal of Artificial Intelligence for Medical Sciences SP - 97 EP - 102 VL - 2 IS - 1-2 SN - 2666-1470 UR - https://doi.org/10.2991/jaims.d.210617.001 DO - 10.2991/jaims.d.210617.001 ID - Yayuan2021 ER -
