Research on Construction of Knowledge Graph of Intestinal Cells
 , Ling Zhang2,
, Ling Zhang2,  , Wei Qu2, Chong Teng3, Dan Xie1, *,
, Wei Qu2, Chong Teng3, Dan Xie1, *, 
- DOI
- 10.2991/jaims.d.200902.001How to use a DOI?
- Keywords
- Intestinal cells; Knowledge graph; Knowledge base; SPARQL query
- Abstract
Intestinal cells play a significant role in human physiological metabolism, immune protection, and development and control of nervous system diseases. With the flourishing development of artificial intelligence technology and arrival of intestinal cellular research enthusiasm, how to obtain knowledge of intestinal cells from a sea of medical literature and realize knowledge visualization has brought great challenges to medical researchers. At present, knowledge graph of intestinal cells has not been studied. In this paper, we present two processes to construct a knowledge graph of intestinal cells: conceptual layer design and instance layer construction. Conceptual layer design defines data model of knowledge graph. In the process of constructing instance layer, cells are regarded as basic conceptual unit to systematically sort out intestinal cell terminology, acquired data is pre-processed, and then mapped into knowledge graph to construct a knowledge base. We provide several SPARQL query cases, by which medical workers can efficiently obtain knowledge they require from the knowledge base, thereby better serving medical researches.
- Copyright
- © 2020 The Authors. Published by Atlantis Press B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
A large number of cells in the intestine play a vital part in the host physiological metabolism, immune protection, and development and control of nervous system diseases. There is a symbiotic relationship between intestinal cells and their hosts. They can work together to maintain balance of human body. Imbalance of gut ecosystem could bring about heavy health damage of hosts, and dietary habits and lifestyle of the hosts, in turn, could also affect diversity of intestinal cells [1]. Intestinal cells have gradually become one of the hot research subjects in the medical domain in recent years. However, owing to an increasing number of medical documents and complex medical concepts, it is troublesome for some researchers to concatenate all medical knowledge that they have learned. With the advent of big data era, the workload is high that effectively storing knowledge extracted from a sea of medical literature and realizing knowledge fusion and visualization by virtue of traditional database. The rapid development of artificial intelligence technology provides people with a new method: knowledge graph. It can effectively realize knowledge structured storage and graph visualization.
Google officially put forward the concept of knowledge graph in 2012, an auxiliary knowledge base used by Google to enhance its search engine functions [2]. The birth of knowledge graph is based on a series of early concepts, including semantic network proposed in 1960 [3], and subsequent ontology [4], Web, Semantic Web [5], Linked Data [6]. Knowledge graph is composed of nodes and edges. The former can be entities or concepts, and the latter can be attributes of entities or relationships among entities. The early idea of knowledge graph comes from the Semantic Web [7]. Figure 1 shows the framework of knowledge graph. It can be seen from the diagram that knowledge graph is a computable and inferable model between knowledge, aiming to identify and extract entities and complex relationships among entities from data with varying degrees of structure. During knowledge graph construction, acquired knowledge is standardized and mapped into knowledge graph. Knowledge base can realize knowledge reasoning and hence continuously improve quality of knowledge graph. Since knowledge graph was come up, a lot of knowledge base projects have emerged at home and abroad. Common large-scale knowledge base projects abroad include ConceptNet, Freebase, DBpedia, BabelNet, Yago, etc. [3]. In addition, there are many large-scale open knowledge graph platforms in Chinese domain [3], such as OpenKG, XLore of Tsinghua University, Zhishi.me of Shanghai Jiaotong University, Belief-Engine of Institute of Automation, Chinese Academy of Sciences. Besides, researchers have also developed many knowledge graphs in the medical domain. However, there is no research to construct knowledge graph of intestinal cells at present. The purpose of this study is to use various information of intestinal cells to build a knowledge graph of intestinal cells, such as regulatory cytokines of intestinal cells, signaling pathways, and functions, etc. This will facilitate medical researchers to query related knowledge, so that knowledge of intestinal cells can be organically connected among a great deal of medical literature.
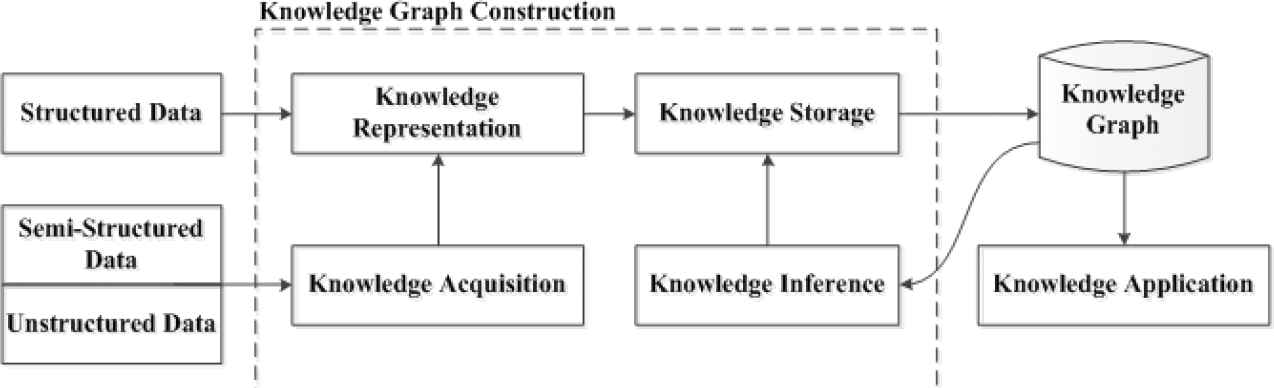
The framework of knowledge graph.
The primary contributions of this paper are as follows. (i) We constructed a knowledge graph of intestinal cells and fused various information of intestinal cells to form a intestinal cellular knowledge base. It is convenient for scholars in intestinal domain to study mechanism of action that different cytokines regulate different signal pathways, thereby better serving medical researches. (ii) Apply knowledge graph of intestinal cells to knowledge retrieval, and use SPARQL query statements to retrieve triples. Thus we gave several SPARQL query cases to enable more researchers to grasp fundamental grammar of SPARQL. In this way, medical researchers can not only obtain knowledge they need from knowledge base, but also connect complex medical concepts among different references.
2. RELATED WORK
Researchers have already developed many professional knowledge graphs in their respective fields since knowledge graph was proposed. As one of the research hotspots in the world today, medical domain is naturally no exception. In this section, we introduce projects for knowledge graphs in the medical domain.
Well-known medical knowledge bases contain UMLS [8], SNOMED-CT [9], ICD-10 [10]. The Unified Medical Language System (UMLS) is a biomedical knowledge base of developed by the US National Library of Medicine. UMLS integrated substantial biomedical concepts that can be linked to external resources and relationships among concepts. The systematized nomenclature of medicine clinical terms (SNOMED-CT) is an international clinical reference terminology. SNOMED-CT facilitates semantic interoperability by providing standardized medical concepts. SNOMED-CT has the potential to improve data quality by supporting data reasoning. The International Classification of Diseases, Tenth Revision (ICD-10) is a disease classification system designed to promote international comparability in the collection, classification and presentation of diseases. It inherited the classification structure and coding rules in the applicable version of the ICD published by the World Health Organization, and converted the reported conditions into medical codes. Recently, experts from abroad have focused on researches over algorithms or models to optimize key technologies for constructing medical knowledge graph. Another direction is to build personalized healthcare knowledge graphs. Andres Duque et al. proposed a graph-based unsupervised technique to address word sense disambiguation in biomedical documents [11]. They used abstracts downloaded from PubMed to build knowledge graph, and used Personalized PageRank algorithm to perform entity disambiguation. Tsendsuren Munkhdalai et al. showed support vector machines model outperformed other model in extracting medical entities and relationships among them from electronic health records via experiments [12]. Amelie Gyrard et al. designed a personalized healthcare knowledge graph for contextualizing information on health diseases [13]. With the help of subject matter experts, Anna Lisa Gentile et al. presented a functional pipeline to maintain up-to-date knowledge resources extracted from Medication Package Inserts based on personalized knowledge graph [14]. In addition, researches on medical knowledge graph also attracted attention from domestic experts. Lirong Jia et al. used Chinese medicine data from Chinese medical language system to construct a knowledge graph of traditional Chinese medicine [15]. Yingying Zhang combined tongue image diagnosis with knowledge graph, and invented a tongue image diagnosis and treatment system for traditional Chinese medicine [16]. Chaoning Cui built a painful perception knowledge graph by painful perception ontology defined and provided several SPARQL query cases based on painful information of patients [17]. Yani Jin et al. divided medical examination knowledge graph construction into two parts: conceptual layer and instance layer, and applied knowledge graph to medical examination knowledge query [18]. Meijie Yang et al. obtained diabetes information from different data sources with web crawler, constructed a diabetes knowledge graph, and implemented question answering system to assist clinicians in making medical decisions [19]. Highly specialized medical knowledge determines that disease field often includes many independent specialized sub-fields, and it is difficult to construct a knowledge graph that can embrace all disease information. Ming Sheng et al. proposed an extensible framework of knowledge graph, which can extend new disease knowledge graph in the existing disease-oriented knowledge graph [20]. Due to significance of medicine to human beings and particularity of medical knowledge, Kaiqi Yuan et al. believed that medical knowledge graph will become a cutting-edge issue for future research [21].
After investigation of domestic and foreign literature reviews, we arrive at the conclusion that although medical field is receiving much attention from experts in knowledge graph, there is no research to integrate knowledge graph with intestinal cells. Inspired by the previous works, in this study, we innovatively propose a knowledge graph of intestinal cells. This can realize structured storage and visual representation of intestinal cellular knowledge and facilitate knowledge query, thereby better serving medical researches.
3. METHODOLOGY
3.1. RDF and RDFS
R. Davis gave a few definitions of knowledge representation: knowledge representation is a substitute indicating an objective entity, or a set of ontological conventions that represent a conceptual model used to describe objective existences [22]. It can be understood that knowledge representation refers to a technique for modeling knowledge in a specific language, scilicet marking objectively existing knowledge with computer symbols to support computers to infer. Nowadays the Internet mostly uses knowledge representation frameworks based on the Semantic Web, such as XML, RDF, and OWL.
Common form of knowledge representation is RDF (Resource Description Framework) triples, (subject, predicate, and object). Subject can be an entity, predicate can be relationships among entities or attributes of entity, and object can be another entity or values of attributes. Taking “Human eosinophils in the gut regulate functions related to cell death through 5-HT2A and 5-HT1/5-HT2 receptor pathways” as an instance, entities and relationships extracted can be rewritten as triples: (Human eosinophils, hasCytokines, 5-HT) and (receptor pathways, regulate, cell death). When rising to the level of the Semantic Web, entity can be represented by URI or URL, and it can be linked to an external resource. Based on a consistent naming scheme, RDF gives us the ability to describe source data in powerful ways and add them to knowledge base. Furthermore, another important value of RDF is to place resources in a global context. The references of the entire knowledge base can be linked to other references, and when citing resources, we can obtain more information about them by querying metadata. Nevertheless, complicated relationships between data made it so untoward for RDF to fully express knowledge that RDF Schema (RDFS) came into being. RDFS added relationships descriptions on the basis of previous RDF. In RDFS, common description keywords of relationships among entities include rdfs:range and rdfs:domain, etc.
3.2. GraphDB and SPARQL
Knowledge graph storage is to save preprocessed and interrelated facts into corresponding knowledge base according to data type. The RDF triples-oriented storage method is triple warehouse, such as GraphDB [23] developed by Ontotext. It implements storage and inference layer API in RDF4J framework, transparent to users. Thus GraphDB is seamlessly connected to RDF4J, users directly accessing GraphDB by means of RDF model and query engine provided by RDF4J. An important feature of GraphDB is to support inference function of triples by virtue of a rule-based inference engine.
RDF supports a declarative query language: SPARQL [24], a query language and data acquisition protocol exclusively for data of RDF triples. As a structured query language, its grammatical format and query form are similar to SQL of relational database. Operations of SPARQL on RDF data mainly include data query, and SPARQL provides four common data query methods: SELECT, CONSTRUCT, ASK, and DESCRIBE. The core processing unit of SPARQL query is triple pattern in query condition. Difference from normal format of RDF triples is that subject, predicate, or object can be variables in triple pattern of SPARQL query. We provided a few typical SPARQL query cases in Section 5. Related medical researchers can study these cases and design appropriate SPARQL statements by modifying variables in querycases.
4. CONSTRUCTION OF KNOWLEDGE GRAPH OF INTESTINAL CELLS
Construction of knowledge graph of intestinal cells can be divided into following two processes: conceptual layer design and instance layer construction. Conceptual layer defines entities, attributes, and relationships among them, emphasizing data representation of entire knowledge graph. Building instance layer includes knowledge acquisition, representation and storage, emphasizing graph realization, and visualization. Therefore, in the process of establishing knowledge graph of intestinal cells, the ontology modeling tool Protégé is first adopted to define data model of knowledge graph so that conceptual layer can be formulated. During instance layer construction, data of intestinal cells is expressed in the form of triples of RDFS, and then facts of triples are imported into GraphDB to constitute knowledge graph of intestinal cells.
4.1. Design Conceptual Layer
The inception phase of constructing knowledge graph of intestinal cells is to design conceptual layer of knowledge graph. That means defining data model of entire knowledge graph and obtaining a macroscopical grasp of overall data pattern of knowledge graph. Then map entities and relationships of intestinal cells into conceptual layer on the basis of previous procedure to complete instance layer. Consequently, conceptual layer design is pivotal in the process of achieving knowledge graph of intestinal cells. In this paper, conceptual layer design of knowledge graph was ultimately determined as follows.
Firstly, definition of intestinal cellular entities is given based on fact records contained in knowledge graph.
Definition 1.
Intestinal cellular entity Cell: Intestinal cellular entity Cell is a medical entity that is uniquely identified by CUI in knowledge graph. Cell entity contains five conceptual entities unfolding in the form of a tree structure: CUI entity, NCI entity, References entity, Cytokines entity, and Pathways entity. For instance, if the References entity serves as the root node, the lower level should comprise the following four conceptual nodes: PubmedID node, DOI node, Date node and Level node. Correspondingly, the three sub-nodes of Pathways entity are: Kegg node, Function node and Description node.
Secondly, design the following sorts of entity relationships based on facts of intestinal cells. (A) SubClassOf indicates that the relationship between two entities is a subordinate relationship between parent class and child class. (B) InstanceOf indicates that the relationship between two entities is a relationship between class and instance. (C) HasReferences indicates the relationship that an intestinal cell is mentioned by a reference. (D) HasCytokines indicates the relationship between an intestinal cell and regulatory cytokines it possesses. (E) HasPathways indicates the relationship of cytokines regulating a certain signal pathway.
According to structural principle of conceptual layer designed above, conceptual layer structure diagram of intestinal cells knowledge graph charted by the ontology modeling tool Protégé is shown in Figure 2.
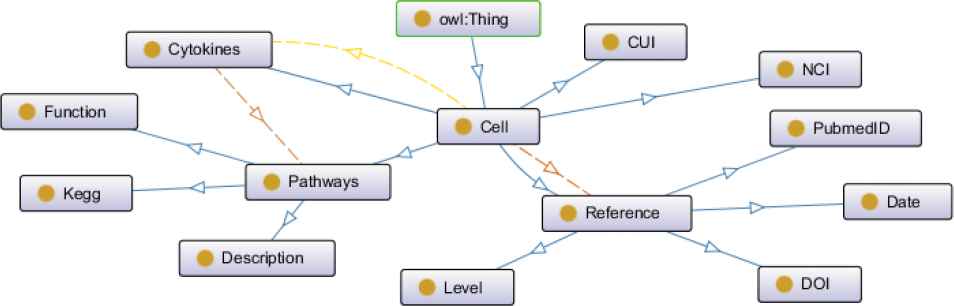
Conceptual layer structure diagram of intestinal cells knowledge graph. The nodes shown in yellow circles indicate various entities of knowledge graph. The edges in different colors indicate different relationships. The blue edges: subClassOf, the yellow edges: hasCytokines, the dark brown edges: hasReferences, the light brown edges: hasPathways.
4.2. Construct Instance Layer
During constructing instance layer, we introduce data acquisition source in the first place, and then data of intestinal cells is expressed in the form of triples. After integrating local triples with SNOMED-CT, facts of intestinal cells are imported into the triples warehouse GraphDB to build knowledge graph of intestinal cells in accordance with conceptual layer structure mentioned above.
Data acquisition Physiological activity of intestinal cells is closely related with human depression, nervous system diseases, autism, anxiety, etc. It is essential for recognizing occurrence and development of mental illnesses and optimizing treatment measures for patients to understand interactions and mechanisms between intestinal cells and psychiatric diseases. For this purpose, we have retrieved 169 relevant papers in PubMed based on three keywords: mental illness, intestinal cell, and review. Study titles, abstracts, and keywords of these references to furtherly determine whether it was worth reading main body according to their relevance to intestinal cells and mental diseases. When reading texts of valuable documents, 47 qualified documents were selected on the basis of inclusion and exclusion criteria of articles shown in Table 1. Papers that were included are content involves in mental illness and intestinal cells. Papers that were published before 2000 and belonged to conference summaries or case reports were excluded. However, although the 47 articles all met abovementioned conditions, it cannot be confirmed that it was intestinal cells that affected mental illnesses of patients. Therefore, 31 references were finally determined based on deepgoing analysis of correlation between mental diseases and intestinal cells. We extracted regulatory cytokines, signaling pathways, and corresponding functions of intestinal cells from these references.
| Inclusion criteria | The research subject of the paper suffered from mental illness |
| The research content of the paper contained intestinal cells. | |
| Exclusion criteria | The paper was published before 2000. |
| The paper type is a conference summary or a case report. |
Inclusion and exclusion criteria of articles.
Knowledge representation based on RDFS Each piece of information is well recorded with cell as the fundamental conceptual unit. Knowledge representation method based on RDFS is used to standardize data of intestinal cells, which can not only effectively solve the problem of non-standard medical data formats, but also achieve structuralization of medical data with discrete computer symbols and facilitate knowledge storage. In this section, taking conceptual layer proposed above as the standard, original data of intestinal cells will be compiled into RDFS triples. To describe these concepts better, we named knowledge graph of intestinal cells as “ICKG”. First of all, part of codes that define entity classes, attributes, and relationships are as follows:
<http://intestines_cell.com/ICKG#reference> a rdfs:Class ;
rdfs:comment ”describe which paper the cell comes from” ;
rdfs:label ”reference”@en .
<http://intestines_cell.com/ICKG#cell> a rdfs:Class ;
rdfs:comment ”describe cell concluded from a paper” ;
rdfs:label ”cell”@en .
<http://intestines_cell.com/ICKG#relationship> a rdfs:Class ;
rdfs:comment ”describe relationship between intestinal cells and mental disorder in references” ;
rdfs:label ”relationship”@en .
<http://intestines_cell.com/ICKG#hasReference> a owl:ObjectProperty;
owl:inverseOf <http://intestines_cell.com/ICKG#hasCell> ;
rdfs:domain <http://intestines_cell.com/ICKG#cell>,
<http://intestines_cell.com/ICKG#relationship> ;
rdfs:range <http://intestines_cell.com/ICKG#reference> ;
rdfs:comment ”this cell is from which paper”@en .
Secondly, taking the second record in original data as an instance, part of codes that information of mouse dendritic cells was rewritten into RDFS triples are as follows.
ickg:ref1_cell2 a ickg:cell ;
ickg:hasCell_name ”mouse dendritic cells” ;
ickg:hasCUI ”C0011306” ;
ickg:hasNCI ”C12583” ;
ickg:hasReference ickg:ref1 ;
rdfs:label ”mouse dendritic cells”@en .
Knowledge storage based on GraphDB GraphDB is a triples-oriented knowledge base and involves rule-based reasoning function. After all data of intestinal cells is expressed in the form of RDF Schema, facts of triples can be stored in the GraphDB. GraphDB also supports knowledge graph visualization and SPARQL query. SNOMED-CT is an international clinical reference terminology that provides standardized medical concepts. We designed a semantic mapping structure and integrated 2443 local facts of triples into concepts of SNOMED-CT. After importing processed triples into GraphDB, the whole knowledge base contains 160253 facts altogether with 546 new records inferred from existing triples.
Knowledge graph visualization In order to demonstrate a visual graph by facts in knowledge base, we chose CONSTRUCT query statements to make node configuration on knowledge graph. That included configuring starting nodes and expansion nodes. Having accomplishing configuration, conceptual entities in knowledge base were mapped into a visual knowledge graph, and part of the graphical result is shown in Figure 3.
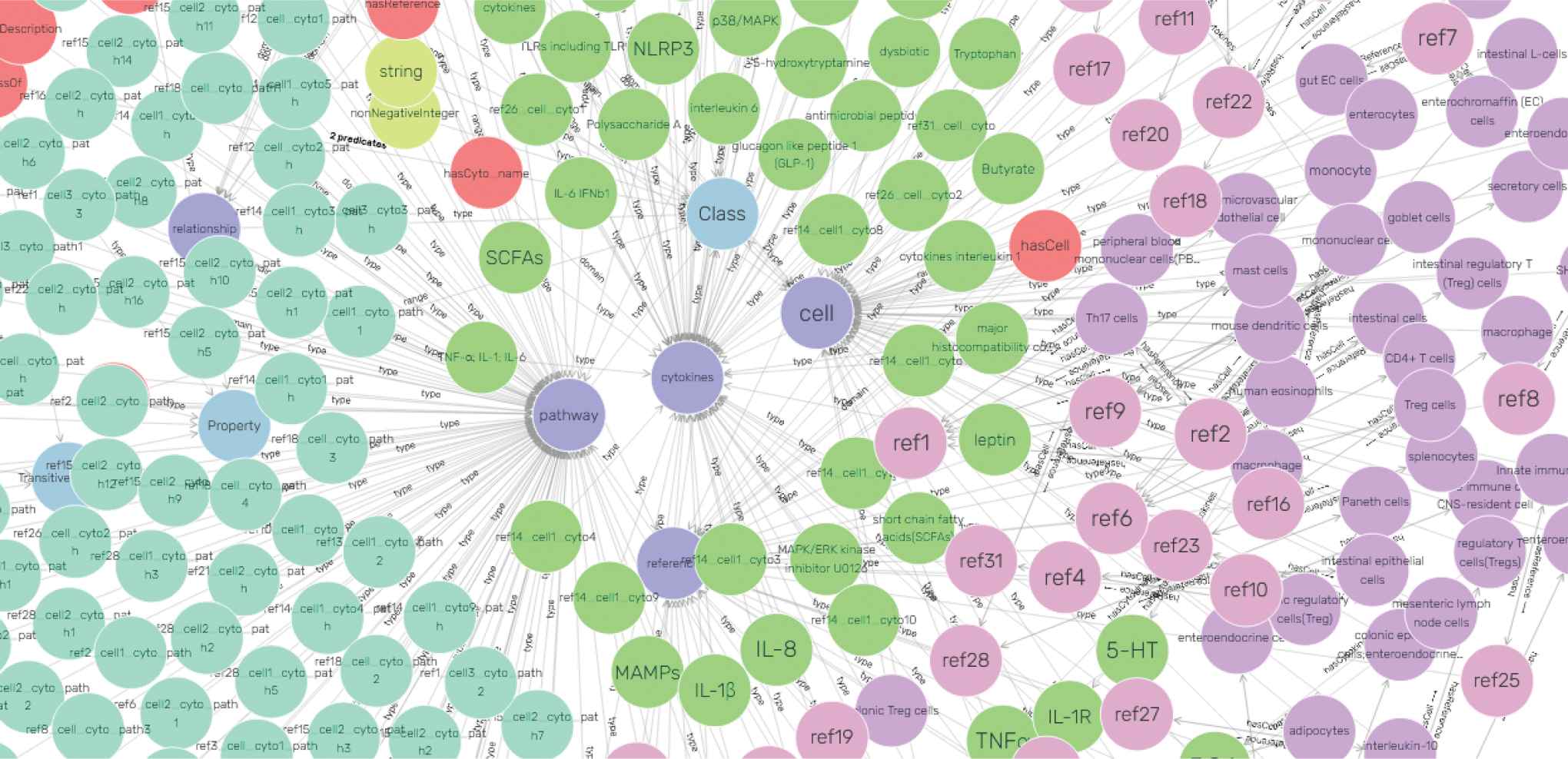
The visualization of intestinal cells knowledge graph. Entities are shown as circles in different colors, such as green cytokines nodes, pink reference nodes, and more. Relationships among entities are represented by edges with arrows.
5. APPLICATION OF KNOWLEDGE GRAPH OF INTESTINAL CELLS
For well-constructed intestinal cells knowledge graph, its quality assessment, namely verifying value and effectiveness of knowledge graph, can be performed through knowledge graph application. In this section, knowledge graph of intestinal cells is applied to knowledge retrieval based on SPARQL sentence. We provided several query cases used to obtain required knowledge from knowledge base. So relevant medical researchers can use for reference to design appropriate query statements.
5.1. Query Case One
Cell entity is considered as fundamental structure unit of intestinal cells knowledge graph to store knowledge. Knowledge graph connects nodes with a graph structure, thereby concatenating various information of intestinal cells among disparate documents. In gut knowledge base, use following SPARQL statement to query all intestinal cells and their concept unique identifiers mentioned in two specified documents. The query generated 7 results shown in Table 2. That means that the two references refer to 7 intestinal cells relevant to mental disorder: human eosinophils, mouse dendritic cells, brain microvascular endothelial cell, macrophage, mast cells, intestinal regulatory T (Treg) cells and Th17 cells. The SPARQL query code showing as follows:
PREFIX …
SELECT DISTINCT ?refN ?cellN ?cui
WHERE {
{?ref1 ickg:hasPMID ”23336044” ;
ickg:hasRef_name ?refN ;
ickg:hasCell ?c .
?c ickg:hasReference ickg:ref1 ;
ickg:hasCell_name ?cellN ;
ickg:hasCUI ?cui .}
UNION
{?ref2 ickg:hasPMID ”28601415” ;
ickg:hasRef_name ?refN ;
ickg:hasCell ?c .
?c ickg:hasReference ickg:ref2 ;
ickg:hasCell_name ?cellN ;
ickg:hasCUI ?cui .}
}
| refN | cellN | cui |
|---|---|---|
| Nicole L, et al. ACS Chemical Neuroscience (2013) 46-48 | Human eosinophils | C0014467 |
| Nicole L, et al. ACS Chemical Neuroscience (2013) 46-48 | Mouse dendritic cells | C0011306 |
| Nicole L, et al. ACS Chemical Neuroscience (2013) 46-48 | BME cell | C0225336 |
| Nicole L, et al. ACS Chemical Neuroscience (2013) 46-48 | Macrophage | C0024432 |
| Nicole L, et al. ACS Chemical Neuroscience (2013) 46-48 | Mast cells | C0014467 |
| Hartmut Wekerle, et al. Trends in Immunology (2017) | Intestinal regulatory T cells | C1267822 |
| Hartmut Wekerle,et al. Trends in Immunology (2017) | Th17 cells | C2936411 |
The result of query case one.
5.2. Query Case Two
Inquiring the regulatory cytokines and signaling pathways of intestinal cells is a great contribution for medical workers to probe into the relationship between intestinal cells and mental illnesses, and the healing of mental diseases with the help of intestinal cells. The function of blood-brain barrier (BBB) below is affected by the permeability of brain microvascular endothelial cells that constitute BBB [25]. Cytokines can increase the permeability of brain microvascular endothelial cells through three stress kinase signaling pathways NFkB, JAK-STAT, and JNK, and then strike a certain balance of BBB. This is essential for maintaining stability of human brain nervous system and studying conception and evolution of psychiatric disorders. The following statements query all signaling pathways regulated by cytokines of brain microvascular endothelial cells and their codes on KEGG. The SPARQL query code is as follows and the result of query is displayed in Table 3.
PREFIX …
SELECT DISTINCT ?sid ?pN ?k
WHERE {
?c ickg:hasCUI ”C0225336” ;
ickg:hasSCID ?s ;
ickg:hasCytokines ?cy .
?s sct:id ?sid ;
sct:hasEnglishLabel ?sN .
?cy rdfs:label ?cN ;
ickg:hasPathway ?p .
?p ickg:hasPath_name ?pN ;
ickg:hasKegg ?k.
FILTER(lang(?cN)='en')
}
| sid | pN | k |
|---|---|---|
| 45709008 | NFkB stress kinase signaling pathways | ickg:map04064–ko04064 |
| 45709008 | JAK-STAT stress kinase signaling pathways | ickg:map04630 |
| 45709008 | JNK stress kinase signaling pathways | ickg:hsa04010–K04440 |
The result of query case two.
5.3. Query Case Three
To some special intestinal cells, they possess multiple regulatory cytokines. And correspondingly, signaling pathways and functions regulated by cytokines are diverse. For instance, suppose that medical researchers want to query all effects of cytokines of enterocytes on the host. The following SPARQL statements answer the questions with 5 results shown in Table 4. P38/MAPK of enterocytes take an effect in reducing colorectal tumorigenesis by increasing phosphorylation of Sp1 and resulting in intranuclear accumulation of p21, while another cytokines of enterocytes, guanylate cyclase C (GC-C), can prevent degradation of tumor suppressor and promote DNA damage repair and genomic stability [26]. The SPARQL query code showing as follows:
PREFIX …
SELECT DISTINCT ?cytName ?pathway ?function
WHERE {
?c ickg:hasCell_name ”enterocytes” ;
ickg:hasCytokines ?cyt .
?cyt ickg:hasCyto_name ?cytName ;
ickg:hasPathway ?pat .
?pat ickg:hasPath_name ?pathway ;
ickg:hasFunction ?function .
}
| cytName | Pathway | Function |
|---|---|---|
| p38/MAPK | Phosphorilation of Sp1 | Reduce colorectal tumorigenesis |
| p38/MAPK | Intranuclear accumulation of p21 | None |
| GC-C | Degradation of tumor suppressor p53 | Prevent degradation |
| GC-C | Degradation of tumor suppressor p53 | Promote DNA damage repair |
| GC-C | Degradation of tumor suppressor p53 | Promote genomic stability |
The result of query case three.
5.4. Query Case Four
Glucagon-like peptide 1 (GLP-1) originating from intestinal L-cells promotes cellular growth and reduces apoptosis through two kegg pathways: map04210 and K02159. Stimulation of the GLP-1 receptor has proved to be highly beneficial in Alzheimer's disease, Parkinson's disease, and peripheral neuropathy [27]. In addition, kegg pathways of GLP-1 also regulate following diseases: autoimmune lymphoproliferative syndromes, hereditary sensory and autonomic neuropathy, OLEDAID, tumor necrosis factor receptor-associated periodic syndrome, X-linked lymphoproliferative syndrome, and colorectal cancer. The following SPARQL query tells researchers which diseases GLP-1 is associated with and CUI of these diseases. The query returns 8 results shown in Table 5, and the query code is as follows:
PREFIX …
SELECT DSITINCT ?cytN ?dN ?cuiD
WHERE {
?c ickg:hasReference ickg:ref8 ;
ickg:hasCytokines ?cyt ;
rdfs:label ?cN .
?cyt rdfs:label ?cytN ;
ickg:hasPathway ?p .
?p ickg:hasKegg ?k .
?k rdfs:label ?kN ;
rdfs:subClassOf ?class ;
ickg:hasDiseases ?d.
?d rdfs:label ?dN ;
umls:cui ?cuiD .
FILTER(?cN=”SH-SY5Y cells”@en && lang(?kN)='en')
}
| cytN | dN | cuiD |
|---|---|---|
| GLP-1 | Alzheimer's disease | C0002395 |
| GLP-1 | Parkinson's disease | C0030567 |
| GLP-1 | Autoimmune lymphoproliferative syndromes | C1328840 |
| GLP-1 | Hereditary sensory and autonomic neuropathy | C0027889 |
| GLP-1 | OLEDAID | C4303737 |
| GLP-1 | Tumor necrosis factor receptor-associated periodic syndrome | C1275126 |
| GLP-1 | X-linked lymphoproliferative syndrome | C0024314 |
| GLP-1 | Colorectal cancer | C4722085 |
The result of query case four.
6. FUTURE WORK
In this section, we introduce limitations of this study and then reflect on future research direction of knowledge graph of intestinal cells. First of all, original data of intestinal cells knowledge graph we constructed in this paper comes from the medical literature. Yet medical facts may be updated with the follow-up of future experiments. Our knowledge graph of intestinal cells thus needs to properly achieve the integration of new triples and old triples.
In addition, users need to master some fundamental SPARQL syntax in the final stage of knowledge graph application. This is difficult for medical researchers who have never had a database learning experience before. Therefore, designing an intelligent question answering system for knowledge graph of intestinal cells is also one of the main research directions of this paper in the future. That will meets query needs in the form of natural language text from medical researchers.
7. CONCLUSION
In this paper, we constructed a knowledge graph of intestinal cells by two processes: conceptual layer design and instance layer construction. Our knowledge graph can effectively store knowledge of intestinal cells extracted from medical references, and realize knowledge fusion and visualization. It is a great contribution for medical researchers in intestinal fields to concatenate complex medical concepts among different documents. That facilitates study on mechanisms of intestinal cytokines through different signaling pathways. Finally, we designed several typical SPARQL query cases to query various information of intestinal cells in knowledge graph application phase. The query results show that medical researchers can efficiently acquire knowledge they require from the knowledge base.
CONFLICT OF INTEREST
The authors declare no competing financial interest. The authors confirm that all funding sources supporting the work and all institutions or people who contributed to the work, but who do not meet the criteria for authorship, are acknowledged. The authors also confirm that all commercial affiliations, stock ownership, equity interests or patent licensing arrangements that could be considered to pose a financial conflict of interest in connection with the work have been disclosed.
AUTHOR CONTRIBUTIONS
All authors have made significant contributions to the manuscript including its conception and design, the analysis of the data and the writing of the manuscript. All authors have reviewed all parts of the manuscript and take responsibility for its content and approve its publication.
ACKNOWLEDGMENTS
This work has been supported by the funding, Research on Traditional Chinese Medicine Modernization of National Key Research and Development Program of the Ministry of Science and Technology in 2019 (2019YFC1712500), as well as by the project, Research on categorizing and demonstrating the characteristic diagnoses and treatment techniques, methods, prescriptions for preventing common diseases with medicines of 15 minority people (2019YFC1712504).
REFERENCES
Cite this article
TY - JOUR AU - Fengfeng He AU - Ling Zhang AU - Wei Qu AU - Chong Teng AU - Dan Xie PY - 2020 DA - 2020/09/24 TI - Research on Construction of Knowledge Graph of Intestinal Cells JO - Journal of Artificial Intelligence for Medical Sciences SP - 15 EP - 22 VL - 1 IS - 1-2 SN - 2666-1470 UR - https://doi.org/10.2991/jaims.d.200902.001 DO - 10.2991/jaims.d.200902.001 ID - He2020 ER -
