Nanosphere's Verigene® Blood Culture Assay to Detect Multidrug-Resistant Gram-Negative Bacterial Outbreak: A Prospective Study on 79 Hematological Patients in a Country with High Prevalence of Antimicrobial Resistance
Equal contribution
- DOI
- 10.2991/chi.d.190321.001How to use a DOI?
- Keywords
- Sepsis; Hematological patients; Molecular diagnosis; Carbapenem resistance
- Abstract
Infections are a major cause of morbidity and mortality in hematological patients. We prospectively tested a new molecular assay (Verigene®) in 79 consecutive hematological patients, with sepsis by gram-negative bacteria. A total of 82 gram-negative microorganisms were isolated by blood cultures, of which 76 cases were mono-microbial. Considering the bacteria detectable by the system, the concordance with standard blood cultures was 100%. Resistance genes were detected in 20 of the isolates and 100% were concordant with the phenotypic antibiotic resistance. Overall, this new assay correctly identified 66/82 of all the gram-negative pathogens, yielding a general sensitivity of 80.5%, and providing information on genetic antibiotic resistance in a few hours. This new molecular assay could ameliorate patient management, resulting in a more rational use of antibiotics.
- Copyright
- © 2019 International Academy for Clinical Hematology. Publishing services by Atlantis Press International B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Sepsis is a leading cause of mortality and critical illness worldwide [1]. A further challenge is that infection is seldom microbiologically confirmed when empirical antibiotic treatment is started. Even when microbiological tests are definitive, culture-positive “sepsis” is observed in only 30–40% of cases [2]. Thus, new diagnostic tools are necessary to facilitate earlier recognition and more timely management of patients with sepsis or at risk of developing it. Bacterial infections are among the major complications of hematological diseases, particularly after hematopoietic stem cell transplantation (HSCT) [3]. At most cancer centers, bloodstream infections (BSIs) are mainly caused by gram-positive bacteria (about 60%), followed by gram-negative bacteria (GNB; about 25%) and fungi (about 10%) [4]. Fortunately, over the decades, numerous successful strategies have been developed to limit the negative impact of these infections. In fact, with the universal use of prompt empirical antibiotic therapy in case of fever during neutropenia and, in some settings, antibiotic prophylaxis, the fatality rate dropped significantly [5]. Recently, GNB have raised in frequency, accounting for 53–56% of all BSI, with the predominant pathogens being Enterobacteriaceae (Escherichia coli, Klebsiella spp, Enterobacter spp) and nonfermenting GNB (Pseudomonas aeruginosa, Acinetobacter baumannii). Reports from cancer centers in various countries increasingly show multidrug resistant (MDR) GNB [6]. Italy has registered a wide and worrisome spread of carbapenem resistance in the past 10 years [7]. While diagnostic and therapeutic interventions have advanced over the last decades, etiologic shifts among bacteria and their growing resistance to antimicrobial therapy have led to increased mortality in hematological patients [7,8].
The Verigene® gram-negative blood culture system (Nanosphere, Northbrook, IL) is a specific test which uses specific bacterial DNA target hybridization and gold nanoparticle probe-based detection. In our institution, a first pilot study evaluated the potential clinical usefulness of this test dedicated to GNB, underlying a high rate of efficacy, and an important gain in time [9]. These results prepared the ground for the application of the Verigene assay in a hematological population.
We evaluated the Verigene system as a molecular approach for early diagnosis of drug resistance in GNB and its potential impact on the current management of hematological patients.
2. METHODS
From June 2014 to May 2016, all blood cultures positive for GNB at the San Raffaele Hematology and Bone Marrow Transplantation Unit were prospectively tested using both the standard techniques, and the Verigene assay [10,11]. Standard blood cultures were processed by a (bioMérieux, USA). The Verigene gram-negative blood culture test (Nanosphere, Northbrook, IL), is a microarray-based, almost fully automated system used to allow bacterial identification and detection of several resistance genes from positive blood cultures. The turnaround time of this test is 2 hours, with a hands-on time of <10 minutes. The test has been approved for clinical use both in the United States and in Europe [9]. The identification panel includes the following species: Escherichia coli, Klebsiella pneumoniae, Klebsiellaoxytoca, Pseudomonas aeruginosa, Serratia marcescens, Acinetobacter spp, Enterobacter spp, Citrobacter spp, Proteus spp. The panel of resistance marker includes the following genes: extended spectrum beta-lactamase (CTX-M), Klebsiella pneumoniae carbapenemase (KPC), new-Delhi metallo-beta-lactamase (NDM), imipenem-resistant metallo-beta-lactamase (IMP), Verona integrin-encoded carbapenem-resistant metallo-beta-lactamase (VIM), Oxacillin-resistant beta-lactamase (OXA). The laboratory has been set up to perform daily tests on all positive blood cultures, between 8 am and 5 pm during the week and between 8 am and 1 pm over the weekend. All patients received supportive care and prophylaxis against opportunistic infections in accordance with the institutional guidelines, and were screened for enteric colonization by MDR bacteria using rectal swab.
Data are described as raw number (percentage) when reporting frequencies. The concordance rate between the Verigene assay and standard methods was examined. Statistical analyses were performed using IBM SPSS for Windows (IBM Corp. Version 20.0, Armonk, NY).
3. RESULTS
3.1. Study Population
We evaluated the reliability of the Verigene test on all consecutive blood cultures submitted to our center and positive for gram-negative pathogens. These cultures were from 79 hematologic patients undergoing chemotherapy (n = 22), autologous (n = 6), or allogeneic (n = 47) HSCT. Stem cell donors were family haploidentical (n = 22), an HLA identical sibling (n = 7), an unrelated volunteer (n = 16), or cord blood (n = 2). The median age was 52 years (range 20–78) and 56 patients were male. A total of 76 subjects had high-risk hematologic malignancies, with the most common being acute myeloid leukemia (AML) 44.3%. Fifty patients (63.3%) had active disease at sample collection, while 26 (32.9%) were in hematologic remission. Severe neutropenia was present in 61 patients. Eighteen patients had enteric colonization by MDR bacteria.
3.2. Bloodstream Infections
A total of 82 gram-negative microorganisms were isolated by blood cultures; 76 cases were mono-microbial, while 3 were poly-microbial BSI. Considering the bacteria detectable by the system, the concordance with standard blood cultures was 100%. Resistance genes (CTX-M or carbapenemases, as KPC and VIM) were detected in 20 of the isolates (Table 1) and 100% were concordant with the phenotypic antibiotic resistance. We observed only 7.31% (6/82) of phenotypic resistance not detected by the test, belonging to other kinds of resistance mechanisms not related to the genes included in the panel. Overall, the Verigene assay correctly identified 66/82 (Table 1) of all the gram-negative pathogens (9 Klebsiella pneumonia, 41 Escherichia coli, 7 Pseudomonas aeruginosa, 3 Acinetobacter spp, 4 Enterobacter spp, 2 Citrobacter spp), yielding a general sensitivity of 80.5%, which increased to 100% if only the genera and species included in the panel were considered. Sixteen pathogens (19.5%) were not included in the panel, 15 of them were not identified by the system, while there was only one case of misidentification, that is, a S. paucimobilis, identified as Acinetobacter spp. Thus, the positive predictive value was of 98.5%.
| Escherichia coli(n 41) | Klebsiella Pneumoniae (n 9) | Pseudomonas aeruginosa (n 7) | Enterobacter (n 4) | Acinetobacter (n 3) | Citrobacter (n 2) | |
|---|---|---|---|---|---|---|
| Resistance markers | ||||||
| CTX-M | 10 | 5 | ||||
| KPC | 7 | 1 | ||||
| VIM | 1 | |||||
| OXA | ||||||
| Empirical therapy | ||||||
| PTZ | 33 | 6 | 3 | 3 | 2 | 2 |
| MEM | 3 | |||||
| MEM + VAN | ||||||
| Combo anti-CR | 2 | |||||
| Other | 8 | 2 | 1 | 1 | ||
| Therapeutic decision based on Verigene | ||||||
| De-escalation | ||||||
| Stop VAN | 6 | 2 | 1 | |||
| From MEM to PTZ | ||||||
| From combo anti-CR to MEM/PTZ | 2 | |||||
| No change/antibiotic-sparing approaches | 15 | 2 | 3 | 1 | 1 | |
| Escalation | ||||||
| From PTZ to MEM | 25 | 2 | 1 | 1 | 2 | 1 |
| From PTZ/MEM to combo anti-CR | 7 |
Abbreviations: CTX-M: Extended spectrum beta-lactamase, KPC: Klebsiella pneumoniae carbapenemase, VIM: Verona integrin-encoded carbapenem-resistant metallo-beta-lactamase, OXA: Oxacillin-resistant beta-lactamase, PTZ: Piperacillin/tazobactam, MEM: Meropenem, VAN: Vancomycin, CR: Carbapenem-resistant.
Antimicrobial resistance pattern and antibiotic therapy management of the gram-negative bacteria isolated by blood cultures and identified by the Verigene test.
3.3. Clinical Management
We next examined the potential clinical impact of this molecular approach in allogeneic HSCT recipients (n = 47), either in inpatient (n = 39) or outpatient (n = 8) management. In the majority of cases, we were able to start early targeted antibiotic therapy (74.5%), sparing or interrupting nonspecific antimicrobial therapy (63.8% and 38.3%, respectively). In 78.7% of patients, immunosuppressive prophylaxis did not need to be reduced, thanks to a rapid control of sepsis, therefore avoiding the risk of GVHD. Early contact isolation was possible in 36.2% of patients, preventing the spread of infections among patients. Infection-related mortality at 30 days was reported in only 4 patients. An outpatient management was continued in 3/8 hemodynamically stable patients, avoiding hospitalization. The median time from the blood sample collection to the blood culture positivity is 18 hours. According to our organization framework, Verigene yielded results about 20 hours (median) in advance, as compared to the standard blood cultures for antibiotic susceptibility, requiring a median of 2 hours from blood culture positivity for all samples in this study (Fig. 1).
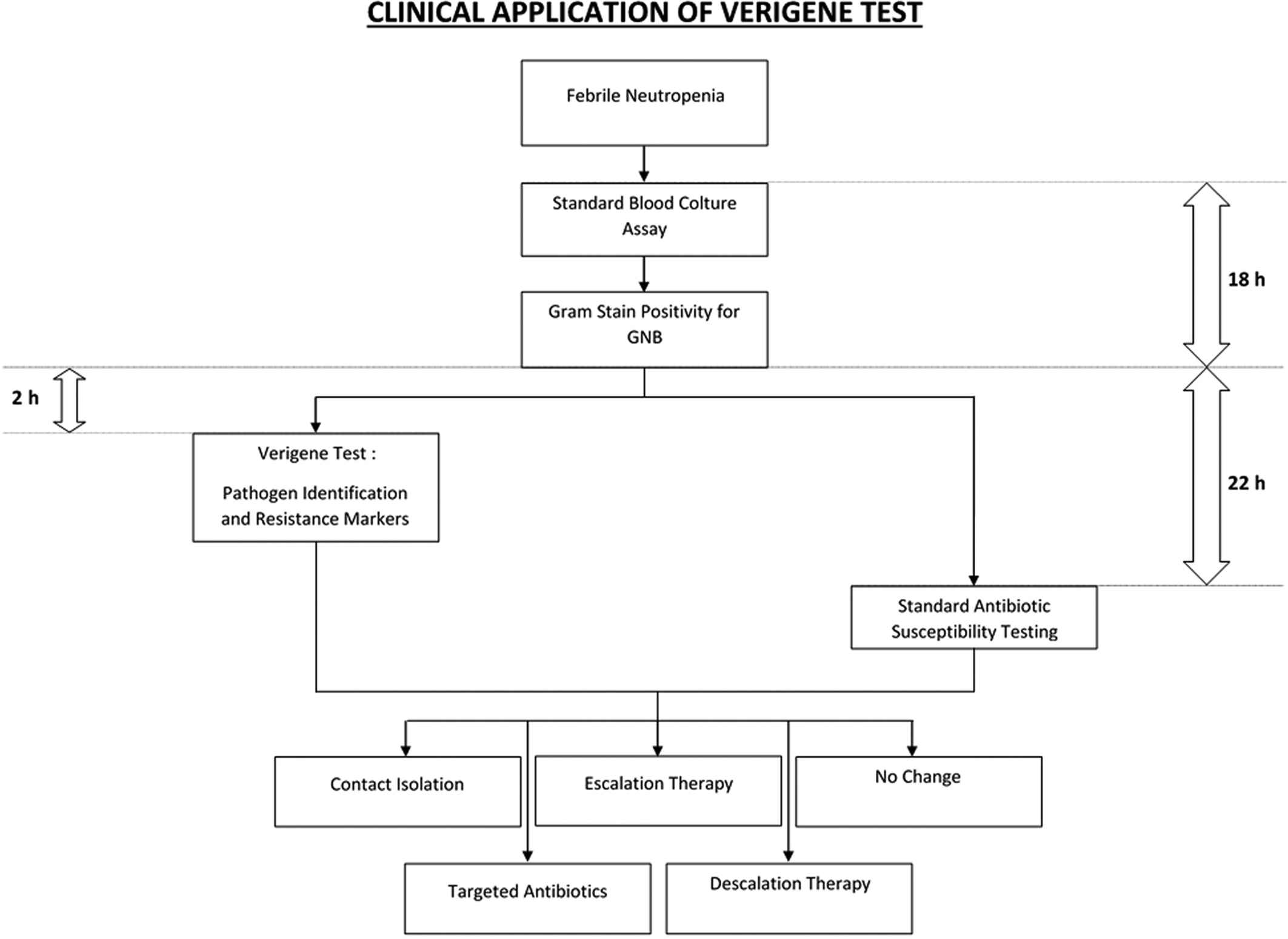
Potential clinical application of the Verigene test and its integration with standard blood culture assay for the management of febrile neutropenia in hematologic patients. The median time from blood sample collection to the blood culture positivity is 18 hours. In our organization, the Verigene test yielded results within a median of 2 hours from blood culture positivity. This represented a gain of a median of 20 hours, as compared to the standard blood cultures with antibiotic susceptibility, which required a median of 22 hours from blood culture positivity.
4. DISCUSSION
A nationwide retrospective survey reported that infection-related mortality due to carbapenem-resistant Klebsiella pneumoniae (CRKp) observed in allo-SCT patients was 64.4% (253). Different studies examined CRKp spread, and active surveillance, together to an accurate transmission prevention, are now broadly recommended [12]. Also in this field, several groups have evaluated the role of molecular approaches. Consequently, we evaluated the clinical impact of the Verigene microarray-based platform on the management of 79 consecutive patients undergoing chemotherapy, autologous or allogeneic HSCT, in an either inpatient or outpatient regimen.
The Verigene test was able to identify genus, species, and genetic resistance determinants for a broad panel of GNB directly from positive blood cultures. We observed a complete concordance with the standard blood culture to detect bacteria included in the Verigene panel and their corresponding phenotypic antibiotic resistance. Importantly, the mean gain in time of Verigene over fast blood cultures was 20 hours in our centre. The clinical application of this assay demonstrated a great influence on patient management. In the majority of cases, we were able to avoid or interrupt nonspecific antimicrobial therapies, reducing unnecessary, inefficient, and/or more toxic antibiotics and their potential impact in favoring antimicrobial resistance. Moreover, in patients colonized by MDR pathogens, the anti-carbapenem-resistant pathogen empirical combination therapy was spared, thanks to the Verigene results that showed the absence of resistance markers. This finding could have a significant impact on the internal guidelines that suggest early initiation of targeted combination therapy in CRKp-colonized patients at the onset of febrile neutropenia. Another important issue addressed in this study was the prompt contact isolation of patients on the basis of an early identification of specific germs or resistance markers, allowing for early prevention of the spread of infections from a patient to another.
In conclusion, the Verigene test represented a significant step forward toward the acceleration and increase of precision of the diagnostic process, providing information on genetic antibiotic resistance in a few hours. This ameliorated patient management, resulting in a more rational use of antibiotics and, ultimately, in improving the outcome of hematologic patients.
CONFLICTS OF INTEREST
All authors: No reported conflicts about this project. After the end of this study, M. Morelli (since September 2017) and A. Forcina (since June 2018) became employees of Novartis; they have not been involved in the final analysis of data.
AUTHORS' CONTRIBUTIONS
R.G., N.M., M.C. and F.C. designed the study. All authors contributed to data collection. R.G., M.C.B. and F.C. analyzed the data. R.G., M.C.B., N.M., M.C. and F.C. wrote the manuscript. All authors have approved the final version of the manuscript.
ACKNOWLEDGMENTS
This study was supported by research grants from Pfizer.
Footnotes
REFERENCES
Cite this article
TY - JOUR AU - Raffaella Greco AU - Maria Chiara Barbanti AU - Nicasio Mancini AU - Laura Infurnari AU - Renée Pasciuta AU - Alessandra Forcina AU - Chiara Oltolini AU - Gabriele Casirati AU - Daniele Mannina AU - Fabio Giglio AU - Carlo Messina AU - Mara Morelli AU - Francesca Lorentino AU - Sara Mastaglio AU - Tommaso Perini AU - Luca Vago AU - Paolo Scarpellini AU - Matteo Giovanni Carrabba AU - Maria Teresa Lupo Stanghellini AU - Sarah Marktel AU - Andrea Assanelli AU - Massimo Bernardi AU - Consuelo Corti AU - Jacopo Peccatori AU - Massimo Clementi AU - Fabio Ciceri PY - 2019 DA - 2019/06/11 TI - Nanosphere's Verigene® Blood Culture Assay to Detect Multidrug-Resistant Gram-Negative Bacterial Outbreak: A Prospective Study on 79 Hematological Patients in a Country with High Prevalence of Antimicrobial Resistance JO - Clinical Hematology International SP - 120 EP - 123 VL - 1 IS - 2 SN - 2590-0048 UR - https://doi.org/10.2991/chi.d.190321.001 DO - 10.2991/chi.d.190321.001 ID - Greco2019 ER -
