Interaction studies on catecholamines to cellular receptors using in silico approach
- DOI
- 10.1016/j.artres.2017.06.001How to use a DOI?
- Keywords
- Catecholamines; Dopamine; Epinephrine; Neurotransmitters; Isoproterenol and nor-epinephrine
- Abstract
Catecholamines are organic compounds derived from amino acids tyrosine and phenylalanine, which acts as neurotransmitters and also functions as hormones in the blood circulation. They bind to plasma proteins and circulate in the blood stream. High levels of catecholamines will cause increase in the heart rate, blood pressure and blood glucose level. These effects are due to binding of catecholamines with adrenergic receptors. Therefore the objective of the current research work is to know the binding affinity of catecholamines with adrenergic receptors through in silico approach. For this study, four catecholamines and three adrenergic receptors were selected for binding analysis. The three natural catecholamines are epinephrine, nor-epinephrine, dopamine and a synthetic catecholamine is Isoproterenol. The three selected receptors are 1GQ4, 3D4S and 1BAK. Binding effect of the four neurotransmitters with the three receptors was studied through in silico analysis using softwares. PATCH DOCK and Z DOCK online servers were used to analyze the docking scores and internal energy was observed by Accelrys Discovery studio. From this study the synthetic catecholamine Isoproterenol showed maximum binding score with all three adrenergic receptors comparing to 3 natural catecholamines. The internal energy of Isoproterenol was found to be 35.18127 kJ/mol. Therefore the study concludes the synthetic catecholamine Isoproterenol has more binding affinity towards beta adrenergic receptors comparing to natural catecholamines. Hence, the current study suggests the usage of synthetic catecholamines will have more binding affinity with adrenergic receptors which could be further analyzed using in vivo study as a future work.
- Copyright
- © 2017 Association for Research into Arterial Structure and Physiology. Published by Elsevier B.V. All rights reserved.
- Open Access
- This is an open access article distributed under the CC BY-NC license.
Background
Catecholamine is a group of amines (nitrogen-contain organic compounds) derived from the amino acid tyrosine and containing a catechol group (aromatic chemical compound consisting of a benzene ring with two hydroxyl groups). Catecholamines are important as neurotransmitters and hormones. They are classified into two major types, natural and synthetic catecholamines. Natural catecholamines are classified into three types they are epinephrine (adrenaline), Norepinephrine (nor adrenaline) and Dopamine. Synthetic catecholamine derivative of epinephrine. The most abundant catecholamines are epinephrine (adrenaline), norepinephrine (noradrenaline) and dopamine, all of which are produced by phenylalanine and tyrosine. Catecholamines as hormone are released by the adrenal glands in situations of stress such as psychological stress or low blood sugar levels.1 The synthesis, function, and degradation of catecholamines reflects the complexity and harmonious coordination in bodily systems. Their synthesis and degradation involve a series of enzymatic steps, and as hormones the catecholamine’s epinephrine, norepinehprine, and dopamine are produced in one part of the body to impact cells in other parts. Various disorders result when this complex coordination is disrupted.
Catechol or pyrocatechol is a benzenediol (aromatic chemical compounds in which two hydroxyl groups are substituted onto a benzene ring) with the formula C6H4 (OH)2. An amine group is a functional group that contains nitrogen as the key atom. Thus, a catecolamine has the distinct structure of a benzene ring with two hydroxyl groups, an intermediate ethyl chain, and a terminal amine group. Some of them are biogenic amines (substances produced by life processes). Catechols can form stable complexes with various di- and trivalent metal ions, the complexes with trivalent ions being the most stable. Catechols can also undergo redox reactions, cycling between catechols, semiquinone radicals and ortho-benzoquinone. The acidity dissociation constants (pKa) for catechol at pH 7: pKa1 – 9.252; pKa2 – 13.0.3 The acidity dissociation constant for the catechol radical can, however, be much lower. The pKa1 of the hydroquinone radical as pH 4.1, and pH 9.85 for hydroquinone.4 Thus, catechol occurs in the non-dissociated form and the catechol radical most probably in the dissociated form at physiological pH. The partitioning coefficients of catechol in an n-octanol water system (Kow) and in a membrane-water system (Kmw) are 7.85 and 4.47 respectively.5 Catechol is not very lipophilic and does not accumulate considerably in membranes or fatty tissues.6 Alkylated or halogenated catechols, however, have higher partitioning coefficients and will therefore accumulate in lipids.
Catecholamines have several functions in the biological system, they help the body respond to stress.7 Catecholamines are water soluble and are 50% bound to plasma proteins, so they circulate in the bloodstream. Two catecholamines, norepinephrine and dopamine, act as neuromodulators in the central nervous system and as hormones in the blood circulation. The catecholamine norepinephrine is a neuromodulator of the peripheral sympathetic nervous system but is also present in the blood (mostly through “spillover” from the synapses of the sympathetic system).8 These hormones cause dilation of the pupils, increase the speed of nerve impulse transmission, widen small air passages in the lungs, and provide additional energy for the body. Norepinephrine causes an increase in blood pressure and epinephrine causes the body’s heart rate to increase.
G protein coupled receptors (GPCRs), also known as seven-transmembrane domain receptors, 7TM receptors, heptahelical receptors,9 serpentine receptor, and G protein-linked receptors (GPLR), constitute a large protein family of receptors that sense molecules outside the cell and activate inside signal transduction pathways and, ultimately, cellular responses.10 The adrenergic receptors are a class of G protein-coupled receptors that are targets of the catecholamines, many cells possess these receptors, and the binding of a catecholamine to the receptor will generally stimulate the sympathetic nervous system.11 The sympathetic nervous system is responsible for the fight-or-flight response, which includes widening the pupils of the eye, mobilizing energy, and diverting blood flow from non-essential organs to skeletal muscle.12
The major focus of this research work done is to identify the binding sites of adrenergic receptors and compare the binding efficiency of different types of catecholamines with adrenergic receptors. This includes the selection of catecholamine compounds, analysis of binding sites of human adrenergic receptor with catecholamines (by in silico studies through discovery studio software)13; to find the compound that shows the best docking score, to check binding pockets and cavities using CASTp and to check for competitive binding using patch dock. The specific reason to choose the adrenergic receptors is mainly to know the exact mechanism of interaction of Isoproternaol (ISPH) compound that has been used in several clinical research studies as a drug to induce experimental myocardial infarction (MI) in experimental animals. Therefore the current study has chosen the adrenergic receptor to know the interaction studies of catecholamines in cellular receptors.
Materials and methods
The binding of ligand molecules (catecholamine), with its respective receptors (Adrenergic receptors) is studied by obtaining the ligand molecules using the PRODRG server and ACCELRYS DISCOVERY STUDIO software. Similar methods were used to obtain Human receptor molecules. Then the docking of receptor and ligand molecules was done using DS LIGANDFIT and DS LIGANDSCORE. The structure of receptor molecules were obtained from protein databases such as RSCB and NCBI. Then the catecholamine compounds were isolated and docked using PRODRG server. Finally, the docking of adrenergic receptors and the catecholamine ligand molecules was done through PACTH dock studies and the results were analysed.
Steps involved in obtaining catecholamine ligand molecules
The steps involved in obtaining Catecholamine molecules, from PRODRG server, were shown in Fig. 1. The other important tools used were Protein Data Bank14 and ACCELRYS DISCOVERY STUDIO.
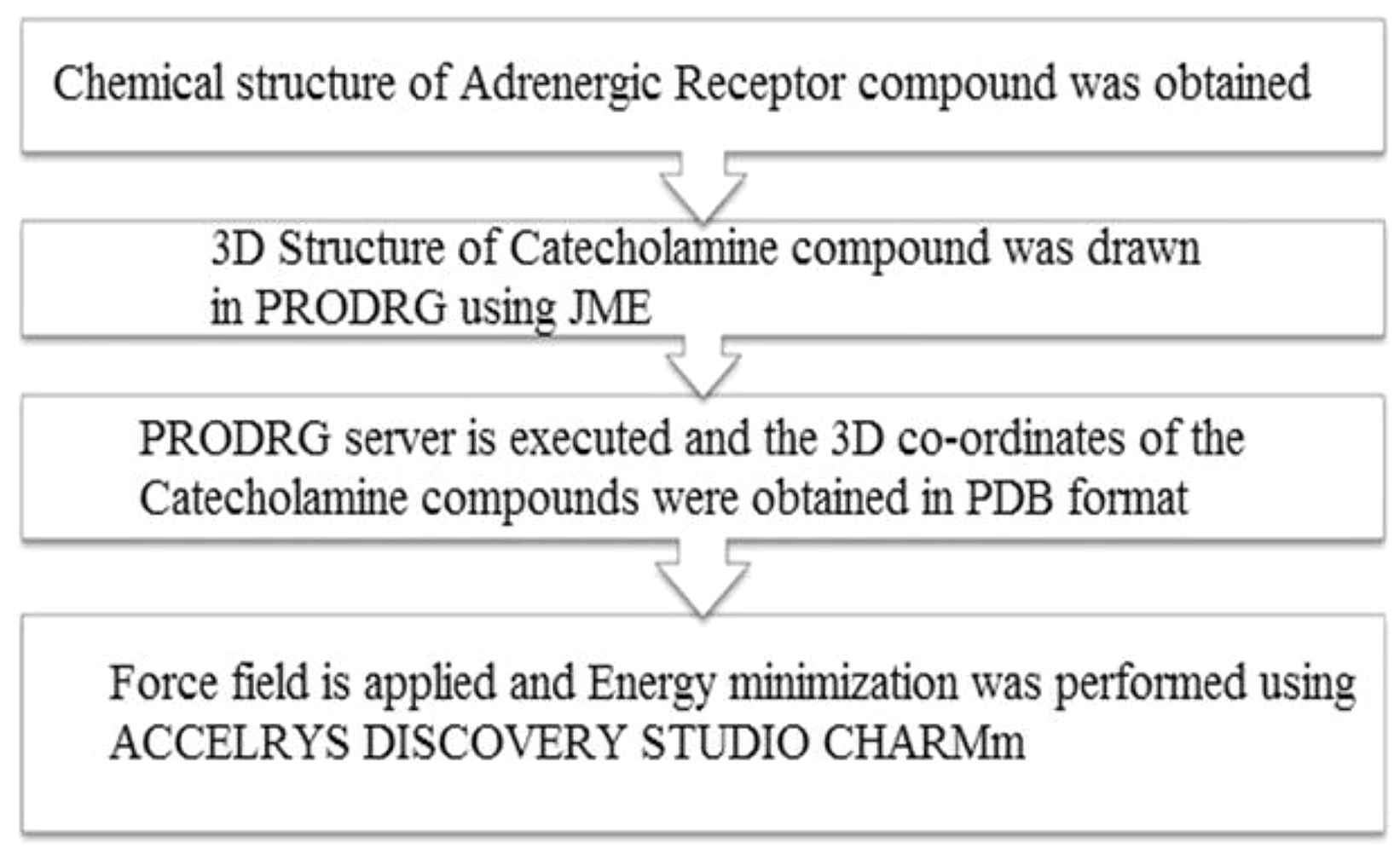
Obtaining catecholamine ligand molecules.
Steps involved in obtaining human receptor molecules
The human receptor molecules were obtained in order to study the binding of catecholamine molecules with the respective adrenergic receptors. The 3D structure of adrenergic receptors were downloaded from RCSB PDB website and were saved as PDB files. Then by further studies, their binding sites were obtained and their interaction with the ligands were studied. Figure 2 shows the processes involved in obtaining the Human receptor molecules. The tools used were RCSB PDB website and ACCELRYS DISCOVERY STUDIO.
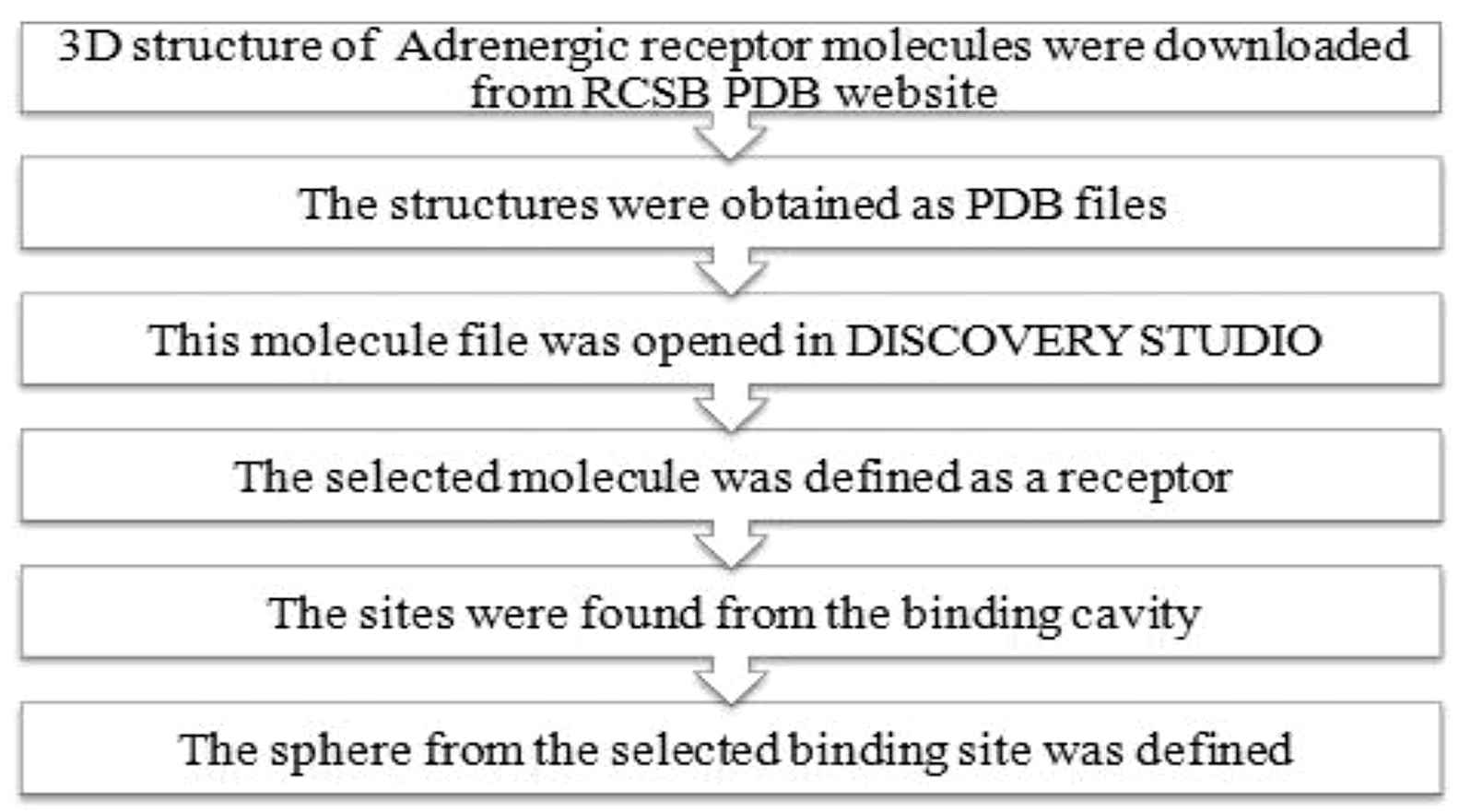
Obtaining human receptor molecules.
Docking of ligand and receptor molecules
Docking of Catecholamines with the adrenergic receptor and the processes involved are shown in Fig. 3. The tools used were DS LIGANDFIT and DS LIGANDSCORE.15,16
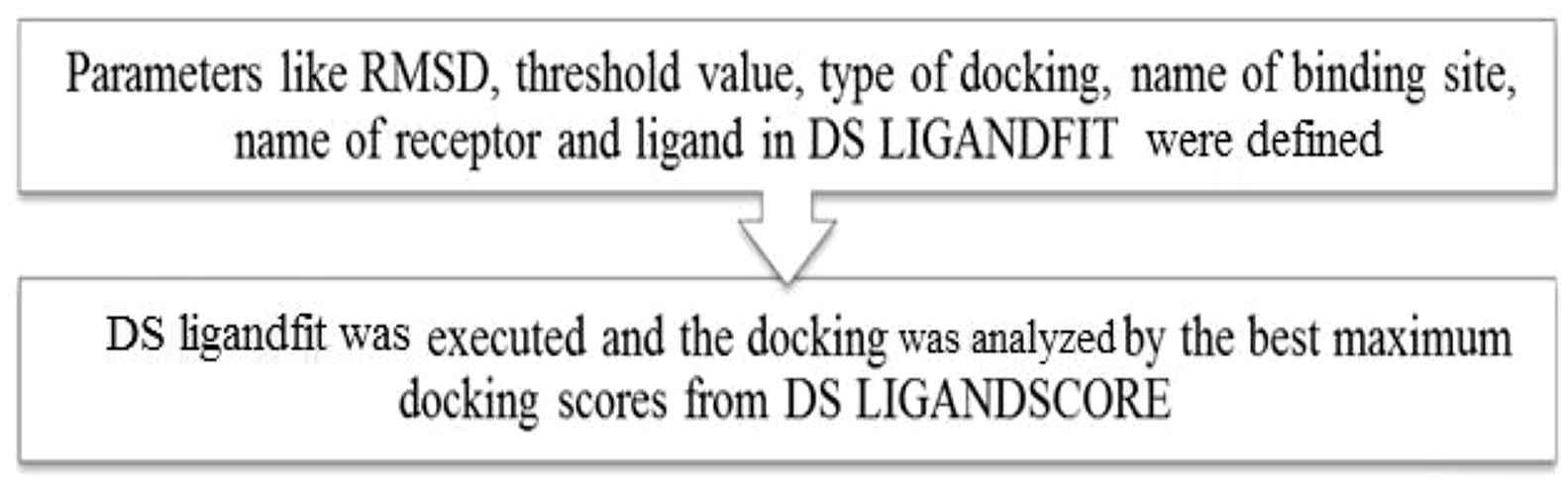
Docking of ligand-receptor molecules.
Structure of required receptor molecules from database: protein data bank (RCSB) and PubChem compound (NCBI)
The structure of the receptor molecules for docking studies were downloaded in the PDB format from protein data bank in the RCSB website. The catecholamine molecules thus taken were used for docking studies. The PubChem Compound database contains validated depiction information provided to describe substances in PubChem substance.17 Structures stored within PubChem compounds were pre-clustered and cross-referenced by identity and similarity groups.18
Obtaining Catecholamine compounds: PRODRG server
The 3-D structure of Catecholamine compounds is not readily available. Hence the 1-D structure was drawn using the JMOL software in the PRODRG server and then the program was executed.5 The resultant file that contains the cartesian co-ordinates with all hydrogen is copied and saved as a PDB format. 3-D structure of Catecholamine compounds thus obtained is used for docking analysis.
Docking with catecholamine compounds
The particular protein molecule data file was opened in Accelrys Discovery studio.19 This opened the protein in a new 3-D window where in the hierarchy view, <cell> was selected and expanded and then the adrenergic receptor molecule was clicked to select. From the tools explorer, Receptor-Ligand interaction was chosen from the dropdown list. Finally under Receptor-Ligand interactions tool panel, define selected molecule as receptor was selected.20 Thus the protein was defined as a receptor for the next step.21 In the binding site tool panel, find sites from receptor cavities option were selected. Then in the hierarchy view, SBD site sphere was selected. SBD site sphere was right clicked and attributes of SBD site sphere was chosen. This opened the sphere object attributes dialog.22 On the dialog, in the radius row the value 9 was entered.23 The radius was then increased to ensure that most of the binding site points are contained within the radius of the sphere. This can be verified visually in graphic view. In the hierarchy view, checkbox beside site 1 was cleared to hide it.
In the files explorer, Catecholamine PDB file was double clicked. In the protocol toolbar, the run button was clicked. The Job completed dialog was displayed once the job got completed. In the Jobs explorer, the completed job was double clicked. This opens the report.html file in the HTML window.24 In the HTML window, in the output files section, the view results link was clicked. This opened the results of the docking which was run.25 The table showed the top score pose alignments for each input ligand. The top score values and internal binding energy values were noted.
From the table obtained, the first row was chosen. Then from the scripts toolbar, show ligand binding sites was chosen. Once the diagram is displayed in graphics view, it was right clicked and labels were chosen. Once the labels window opens, amino acid was chosen from the dropdown list along with an appropriate colour. The interacting amino acids are then labelled onto the diagram. The picture was saved as image files in.bmp format.26
Docking between adrenergic receptor and catecholamine ligand molecules (PACTH DOCK)
The receptor-ligand interaction was done using PACTH DOCK studies. The catecholamine ligand molecules, which were obtained from the previous steps such as from PRODRG server and the receptor molecules obtained from RCSB PDB were interacted with each other, through software docking. The receptor and ligand molecules were opened. The pull down menu “controls” was selected and the clustering RMSD option27 was selected and complex type was changed to default. The RCSB (Research Colloboratory for Structural Bioinformatics) – PDB also provides a variety of tools and resources.28
Results
The above methods were used for the isolation of catecholamine, ligand molecules and the adrenergic receptor molecules through various software tools mentioned above. The results of the studies were recorded and the interaction between the catecholamine molecules and the adrenergic receptors were obtained as mentioned below.
Dock score and internal binding energy
The selected Adrenergic receptor molecules were docked with four catecholamine compounds ie., Epinephrine (adrenaline), Norepinephrine (nor-adrenaline), Dopamine and Isoproterenol. Dock score values and Ligand binding energy values were noted. Dock score value signifies the binding between the receptor and catecholamine compounds. Ligand binding energy value signifies the energy of the binding between the molecules under study.
The Adrenergic receptor molecules taken from study were: 4GBR, 3NY8, 3NY9, 1BAK, 3PDS, 2RH1, 3V5W, 3KRX, 1GQ4, 3D4S, Out of the above ten adrenergic receptor molecules, only three molecules showed positive binding with catecholamine compounds. They were 1BAK, 3D4S and 1GQ4. The following table (Table 1) gives the energy of the adrenergic receptor molecules before minimization and after minimization.
| Name of the human Adrenergic receptor molecule | Energy before minimization using CHARMm (KJ/mol) | Energy after minimization using CHARMm (KJ/mol) |
|---|---|---|
| 3D4S | −26380.19613 | −26184.73572 |
| 1GQ4 | −1218.96223 | −9007.76786 |
| 1BAK | −2883.13271 | −7228.34117 |
Energies of Adrenergic receptor molecules before and after minimization.
The following table (Table 2) gives the details of the energy of the catecholamine compounds before and after minimization:
| Name of the catecholamine compounds | Energy before minimization using CHARMm (KJ/mol) | Energy after minimization using CHARMm (KJ/mol) |
|---|---|---|
| Epinephrine | 35.06690 | −8.08772 |
| Nor epinephrine | 32.59539 | −10.62430 |
| Dopamine | 34.25194 | −2.32628 |
| Isoproterenol | 35.18127 | −5.43410 |
Energies of Catecholamine compounds before and after minimization.
Patch dock
Table 3 shows the score, area ace and transformation results for the ligand molecules that were docked with 1BAK receptor.
| Ligand | Score | Area | Ace | Transformation |
|---|---|---|---|---|
| Epinephrine | 3438 | 385.90 | −110.56 | −0.95 0.60 1.31–26.92 −0.51 4.11 |
| Norepinephrine | 3272 | 349.70 | −115.95 | −1.24 0.31 1.72–25.27 −0.76 4.55 |
| Dopamine | 3046 | 343.80 | −118.87 | 1.95–0.08 2.01–24.07 −1.24 6.12 |
| Isoproterenol | 3724 | 419.20 | −102.00 | 2.50 0.59 0.46–30.10 2.73 1.96 |
Ligand molecules that were docked with 1BAK receptor.
Table 4 shows the score, area ace and transformation results for the ligand molecules that were docked with 1GQ4 receptor.
| Ligand | Score | Area | Ace | Transformation |
|---|---|---|---|---|
| Epinephrine | 2766 | 307.40 | −116.35 | 2.83–0.03 −0.96–8.82 23.55 29.07 |
| Norepinephrine | 2660 | 281.70 | −117.09 | 2.97–0.42 −0.68–8.95 23.90 30.27 |
| Dopamine | 2536 | 271.40 | −139.95 | 3.07–0.07 −0.72–8.82 18.41 28.64 |
| Isoproterenol | 3156 | 344.10 | −105.76 | −0.20–0.66 1.77–2.19 15.49 33.28 |
Ligand molecules that were docked with 1GQ4 receptor.
Table 5 shows the score, area ace and transformation results for the ligand molecules that were docked with 3D4S receptor.
| Ligand | Score | Area | Ace | Transformation |
|---|---|---|---|---|
| Epinephrine | 3322 | 377.20 | −118.17 | −1.06–1.06 −0.77 3.48–0.05 64.55 |
| Norepinephrine | 2942 | 338.00 | −178.01 | 1.72–0.40 0.19–11.97 −4.51 37.74 |
| Dopamine | 3038 | 347.10 | −179.26 | −2.03 0.59–0.55 −10.40–2.63 33.44 |
| Isoproterenol | 3824 | 443.30 | −124.15 | 1.32–0.42 −1.73 5.94 13.86 58.60 |
Ligand molecules that were docked with 3D4S receptor.
All of the results showed above that Isoproterenol only has maximum docking score with the above three receptors.
Accelrys discovery studio
The following table (Table 6) gives the details of the number of positive Binding sites of the Adrenergic receptor molecules with catecholamine compounds.
| S.No. | Name of the Adrenergic receptor molecule | Total number of binding sites generated | Positive docking sites | Negative binding sites |
|---|---|---|---|---|
| 1 | 3D4S | 9 | 5 | 4 |
| 2 | 1GQ4 | 4 | 3 | 1 |
Binding sites of the Receptor molecules.
The following table (Table 7) gives the details of the Dock Score and the Ligand Binding Energy along with other scores of the molecules that showed docking with catecholamine compound.
| Name of the Receptor Molecule | Ligand Score 1 | Ligand Score 2 | PLP1 | PLP2 | Jain | PMF | Dock Score | Ligand Binding Energy |
|---|---|---|---|---|---|---|---|---|
| 3D4S | 3.43 | 4.13 | −35.21 | −40.17 | −0.47 | −61.97 | 50.061 | −1.78 |
| 1GQ4 | 3.02 | 2.95 | 9.87 | 8.78 | −1.01 | 50.04 | 44.54 | −2.14 |
Dock score and Ligand Binding Energy.
Docking poses
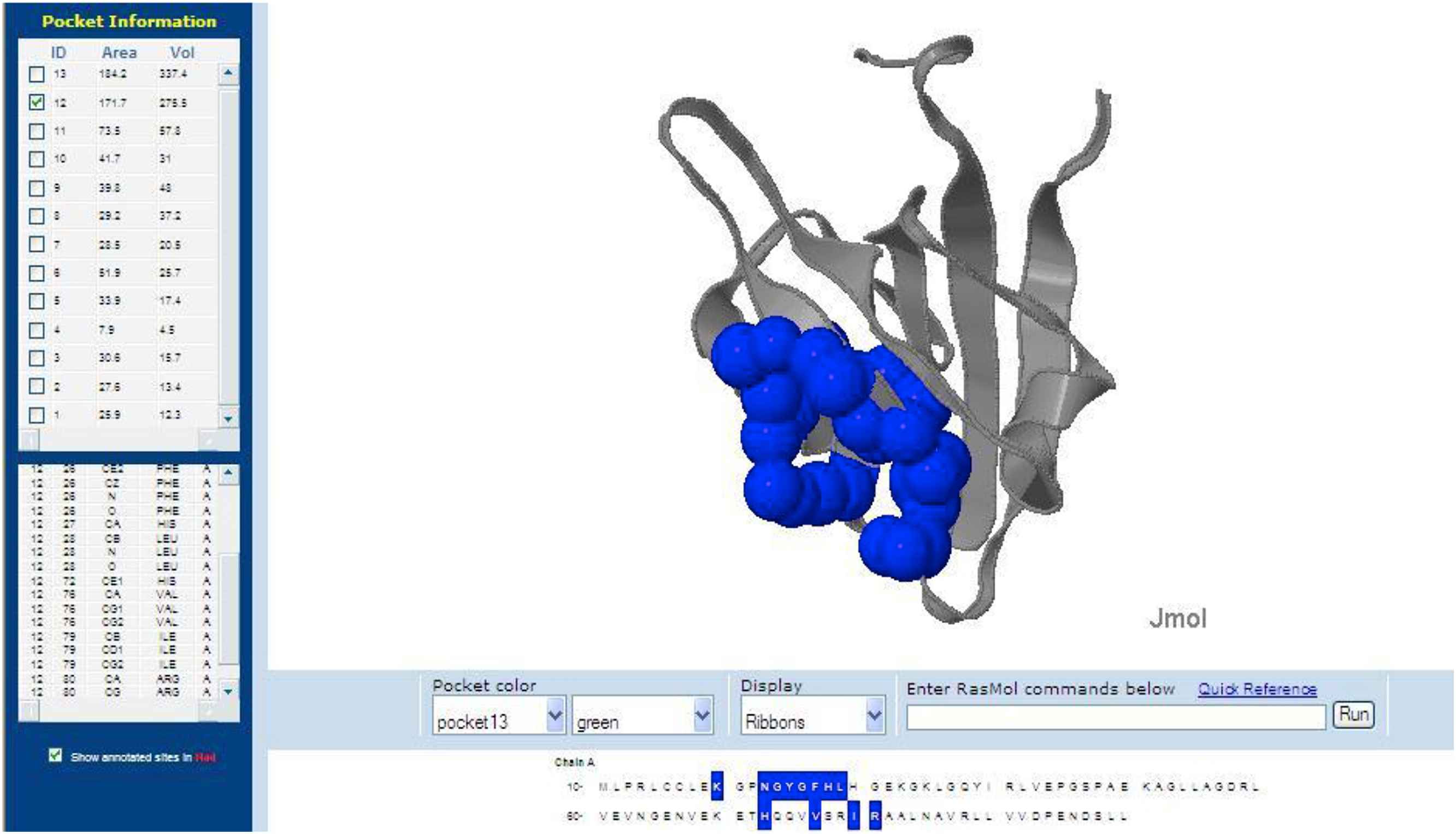
Figure 1 shows the docking pose of 3D4S with Isoproterenol. This reveals the docking of Isoproterenol with the active site of 3D4S with a Dock Score value of 50.061 and Ligand Binding Energy −1.78 kJ/mol. It is noted that the binding occurs only in one region: Binding of carboxyl group of GLU 1011and GLU 1022 with the hydrogen end of Isoproterenol.
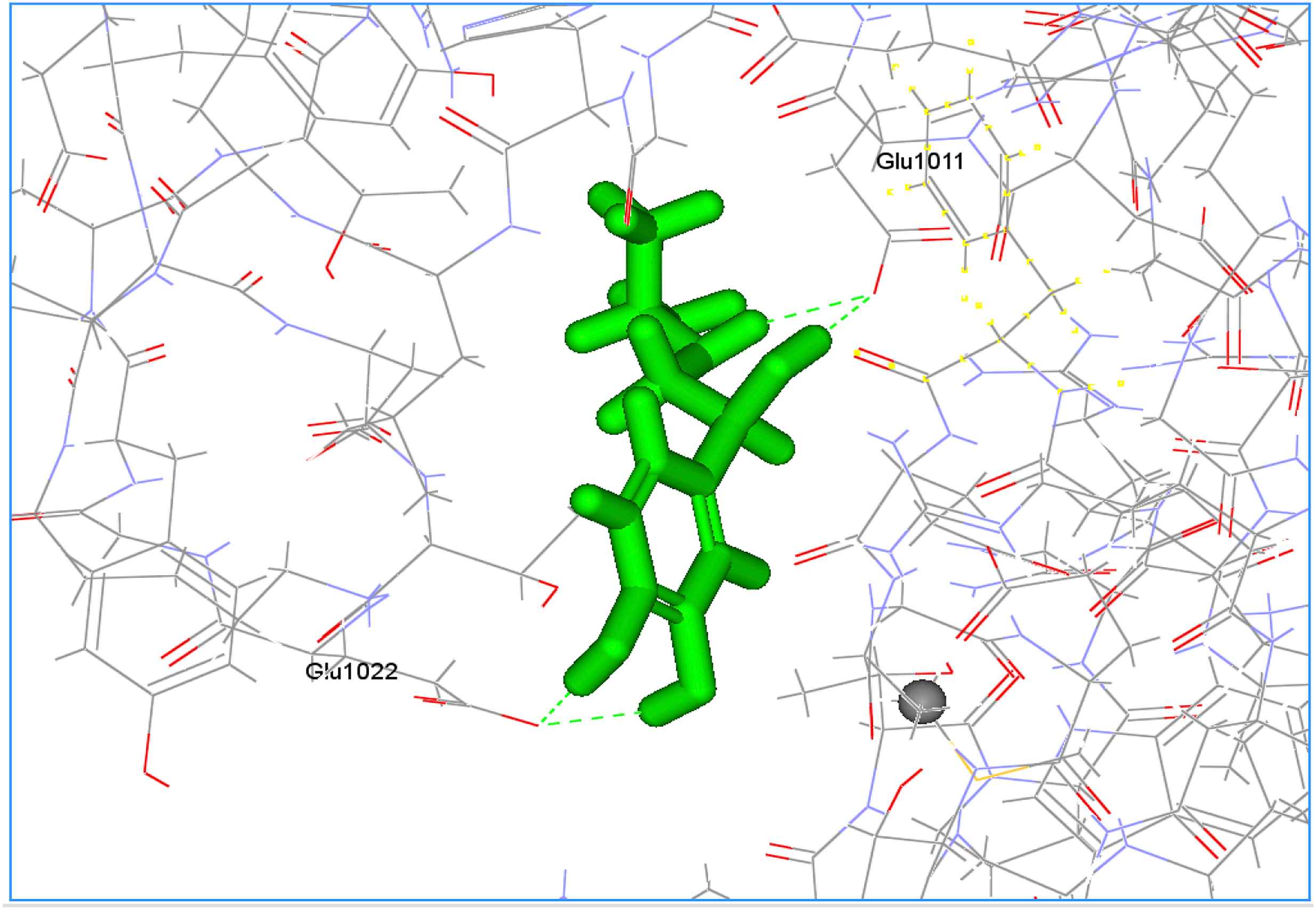
Docking pose of 3D4S with Isoproterenol.
Figure 2 shows the docking pose of 1GQ4 with Isoproterenol. This reveals the docking of Isoproterenol with the active site of 1GQ4 with a Dock Score value of 44.54 and Ligand Binding Energy −2.14 kJ/mol. It is noted that the binding occurs only in one region: Binding of carboxyl group of ARG 80 and CL 1103 with the hydrogen end of Isoproterenol.
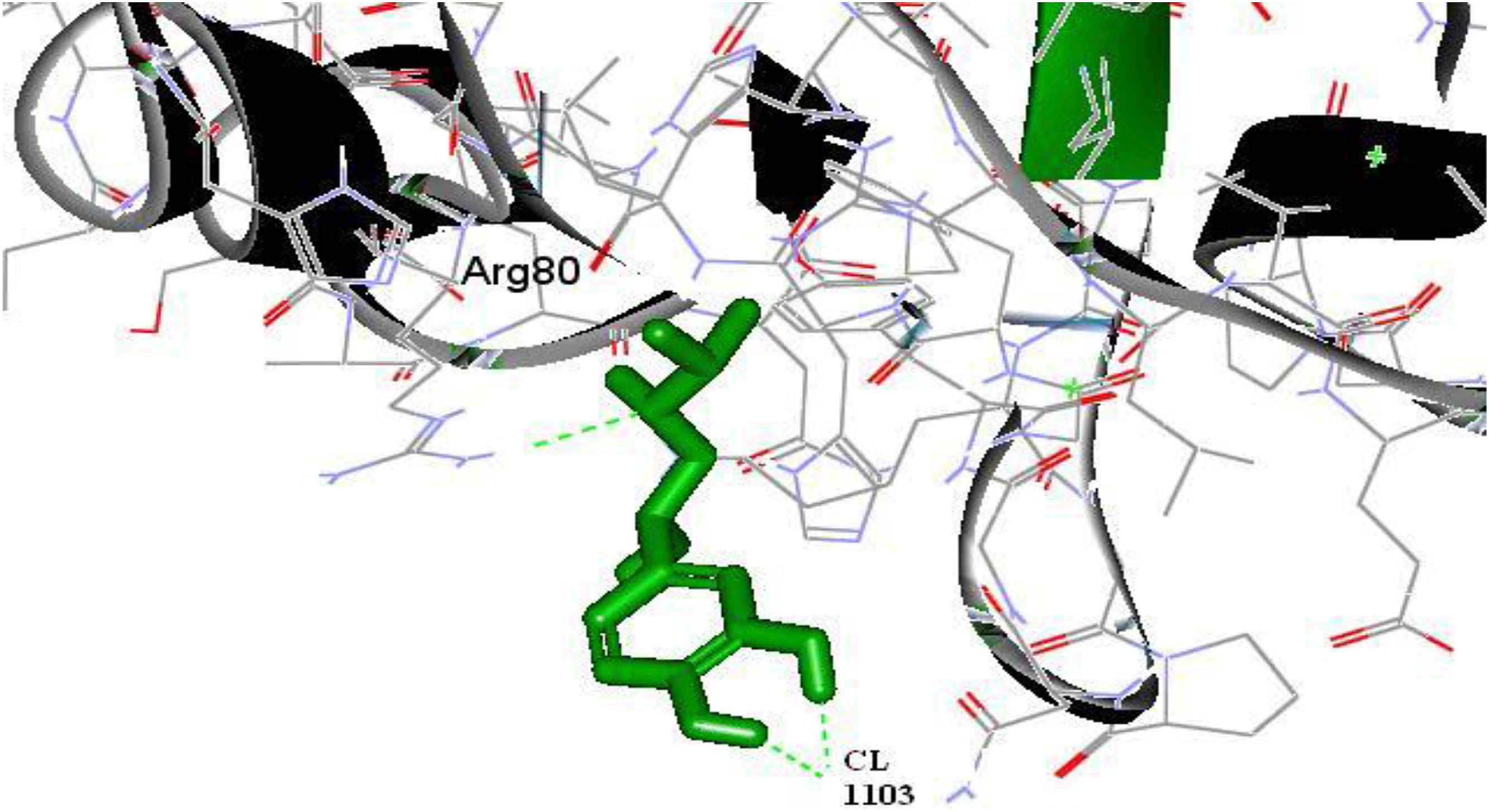
Docking pose of 1GQ4 with Isoproterenol.
Table 8 shows the Catecholamine, the receptor molecule to which it binds and the binding site amino acid data.
| S.No. | Catecholamine | Name of the Receptor Molecule | Binding site AA |
|---|---|---|---|
| 1 | Isoproterenol | 3D4S | A:GLU1011:OE2-Isoproterenol:H16 A:GLU1022:OE2-Isoproterenol:H18 A:GLU1011:OE2-Isoproterenol:H19 |
| 2 | Isoproterenol | 1GQ4 | A:ARG 80:HH21-Isoproterenol:N4 Isoproterenol:H17-A:CL1103:CL Isoproterenol:H18-A:CL1103:CL |
Binding site amino acid.
Discussion
Catecholamines are the organic compounds, secreted from the amino acids.29 It acts as neurotransmitters. Catecholamines cause general physiological changes that prepare the body for physical activity. Some typical effects are increases in heart rate, blood pressure, blood glucose levels, and a general reaction of the sympathetic nervous system.30–32 In the above study, the binding of ligand molecules (catecholamine), with its respective receptors (Adrenergic receptors) was studied by obtaining the ligand molecules using the PRODRG server and ACCELRYS DISCOVERY STUDIO software.33 Similar methods were used to obtain Human receptor molecules. Then the docking of receptor and ligand molecules was done using DS LIGANDFIT and DS LIGANDSCORE. The structure of receptor molecules were obtained from protein databases such as RSCB, NCBI and through Protein Data Bank. Then the catecholamine compounds were isolated and docked using PRODRG server. Finally, the docking of adrenergic receptors and the catecholamine ligand molecules was done through PACTH dock studies and the results were analysed. The results of the analysis were also recorded and explained with structural relevance to the compounds.
Catecholamines were found to be resulting in physiological changes in sympathetic nervous system.34 However in this project it was found that specific type of Catecholamine compound which can bring about the immunological response when bound to human adrenergic receptor molecules using the bioinformatics tool Accelrys discovery studio.35 The Adrenergic receptor molecules which got docked with catecholamines were receptors of 3D4S, 1BAK and 1GQ4.36,37 Thus further in vitro studies can be carried out by using only the molecules which got docked with the human adrenergic receptor molecules.38,39 Thus this will help to reduce the amount of work being done in in vitro.
Conclusion
The Adrenergic receptor molecules with stable interaction with Catecholamines were selected on the basis of internal energy analysed by bioinformatics tools and software. From this it was observed that only three molecules proved positive results with Catecholamines. They were namely 3D4S, 1BAK and 1GQ4. The binding pockets of amino acids are also verified from CASTp server. Out of the four different Catecholamines compounds, Isoproterenol showed best docking scores with the three adrenergic receptor molecules, namely 3D4S, 1BAK and 1GQ4. Thus among the four Catecholamines compounds, Isoproterenol gives best result with the 3D4S molecules. Thus the above results strongly support the neurotransmitter activity of Isoproterenol with the adrenergic receptor molecules. On further analysis and research, the interaction between catecholamine and adrenergic receptors could be used in medical fields to induce experimental myocardial infarction in small animals and to cure severe ailments concerning various cardiovascular, muscular and neurological disorders. It might be also used to Smooth muscle relaxation and Stimulates insulin secretion.
List of abbreviations
- RCSB
Research colloboratory for structural bioinformatics
- NCBI
National center for biological information
- PDB
Protein data bank
- JME
Java mean time editor
- CASTp
Computed atlas of surface topography of proteins
- PLP
Piecewise linear potential
- PMF
Potential of mean force
- DS
Discovery studio
- BLAST
Basic local alignment search tool
- IUPAC
International Union of Pure and Applied Chemistry
- CPR
Cardiopulmonary resuscitation
- ATP
Adenosine triphosphate
- c-AMP
Cyclic- 3′,5′- adenosine monophosphate
- AMPT
Alpha-methyl-p-tyrosine
- GPCRs
G protein coupled receptors
- GPLR
G protein-linked receptors
- TM
Transmembrane
Author’s contributions
K obtained the ligand molecules, results interpretation and also drafted the manuscript. VK obtained Human Receptor Molecules by using similar methods and also docked the receptor and ligand molecules. VS obtained the structure of the receptor molecules and helped to draft the manuscript. PS conceived, participated, coordinated in the designing of the whole study and assisted in tabulating the values. KN supported for editing. All authors read and approved the final manuscript.
Conflict of interest statement
The author(s) declare that they have no conflict of interests in the current study.
Acknowledgment
The authors cordially thank the Management, Secretary, Principal and Head of the Department for granting the permission to use the computer facilities, modeling softwares and the internet facilities for the successful completion of this project.
References
Cite this article
TY - JOUR AU - S. Kalaivannan AU - T. Vinoth kambali AU - S. Prabhu AU - S. Visvanathan AU - N. Karpagam PY - 2017 DA - 2017/06/26 TI - Interaction studies on catecholamines to cellular receptors using in silico approach JO - Artery Research SP - 29 EP - 37 VL - 19 IS - C SN - 1876-4401 UR - https://doi.org/10.1016/j.artres.2017.06.001 DO - 10.1016/j.artres.2017.06.001 ID - Kalaivannan2017 ER -